Waterston RH, et al., including Elnitski L. Initial sequencing and comparative analysis of the mouse genome. Nature, 420: 520-562. 2002. [PubMed]
Gibbs RA, et al., including Elnitski L. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature, 428: 493-521. 2004. [PubMed]
The International Chicken Genome Consortium, Hillier, LW, et. al., including Elnitski L. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature, 432(7018):695-716. 2004. [PubMed]
ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science, 306:636-640. 2004. [PubMed]
Giardine, B., Riemer, C., Hardison, R.C., Burhans, R., Elnitski, L., Shah, P., Zhang, Y., Blankenberg, D., Albert, I., Taylor, J., Miller, W., Kent, W.J., Nekrutenko, A. Galaxy: a platform for interactive large-scale genome analysis. Genome Res, 15:1451-5. 2005. [PubMed]
Elnitski, L., Jin, V.X., Farnham, P.J., Jones, S.J. Locating mammalian transcription factor binding sites: A survey of computational and experimental techniques. Genome Res, 16:1455-1464. 2006. [PubMed]
Petrykowska, H.M, Vockley, C.M., and Elnitski, L. Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Research, 18(8):1238-46. 2008. [PubMed]
Elsik CG, et al. including Elnitski L. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science, 324: 522-528. 2009. [PubMed]
Piontkivska H, Yang MQ, Larkin DM, Lewin HA, Reecy J, Elnitski L. Cross-species mapping of bidirectional promoters enables prediction of unannotated 5' UTRs and identification of species-specific transcripts. BMC Genomics, 10:189. 2009. [PubMed]
Bovine Genome Sequencing and Analysis Consortium, Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, Adelson DL, Eichler EE, Elnitski L, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science, 324:522-8. 2009. [PubMed]
Woolfe A, Mullikin JC, Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome Biol, 11(2):R20. 2010. [PubMed]
Kolbe DL, DeLoia JA, Porter-Gill P, Strange M, Guirguis A, Krivak TC, Brody LC, Elnitski L. Genomic analysis of site-specific DNA methylation patterns in primary epithelial ovarian cancers and endometrial metastases to the ovary. PLoS ONE, 7:e32941. 2012. [PubMed]
Scott, A, Petrykowska HM, Gotea V, Hefferon T, Elnitski L. Functional analysis of synonymous substitutions predicted to affect splicing of the CFTR gene. J. Cystic Fibrosis, 11:511-517. (Top 25 most downloaded paper April-June 2012.) 2012. [PubMed]
The ENCODE Project Consortium, including Elnitski L. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57-74. 2012. [PubMed]
Laflamme K, Owen AN, Devlin EE, Yang MQ, Wong C, Steiner LA, Garrett LJ, Elnitski L, Gallagher PG, Bodine DM. Functional analysis of a novel cis-acting regulatory region within the human ankyrin gene (ANK-1) promoter. Mol Cell Biol, 30:3493-502. 2010. [PubMed]
ENCODE Project Consortium, et. al. including Elnitski, L. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol, 9:e1001046. 2011. [PubMed]
Gotea V, Petrykowska HM, Elnitski L. Bidirectional promoters as important drivers for the emergence of species-specific transcripts. PLoS One, 8:e57323. 2013. [PubMed]
Parker SCJ, Prickett TD Dutton-Regester K, Stitzel ML, Lin JC, Davis S, Simhadri VL, Jha S, Katagiri N, Gotea V, Teer JK, Xiaomu W, Morken MA, Bhanot UK, NISC Comparative Sequencing Program, Chen G, Elnitski LL, Davies MA, Gershenwald JE, Carter H, Karchin R, Robinson W, Robinson S, Rosenberg SA, Francis S. Collins FS, Parmigiani G, Komar AA, Kimchi-Sarfaty C, Hayward N, Margulies EH, and Samuels Y. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci, 110:13481-6. 2013. [PUbMed]
Sánchez-Vega F, Gotea V, Petrykowska HM, Margolin G, Krivak TC, DeLoia JA, Bell DW, Elnitski L. (2013) Recurrent patterns of DNA methylation in the ZNF154, CASP8, and VHL promoters across a wide spectrum of human solid epithelial tumors and cancer cell lines. Epigenetics, 8:1355-72. [PubMed]
Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC. Defining functional DNA elements in the human genome. Proc Natl Acad Sci, 111:6131-8. 2014. [PubMed]
Gotea V, Elnitski L. Ascertaining regions affected by GC-biased gene conversion through weak-to-strong mutational hotspots. Genomics, 103(5-6):349-56. 2014. [PubMed]
Gotea V, Gartner JJ, Qutob N, Elnitski L, Samuels Y. The functional relevance of somatic synonymous mutations in melanoma and other cancers. Pigment Cell Melanoma Res, doi: 10.1111/pcmr.12413. 2015. [PubMed]
Sánchez-Vega F, Gotea V, Margolin G, Elnitski L. (2015) Pan-cancer stratification of solid human epithelial tumors and cancer cell lines reveals commonalities and tissue-specific features of the CpG island methylator phenotype. Epigenetics Chromatin, 8:14. 2015. [PubMed]
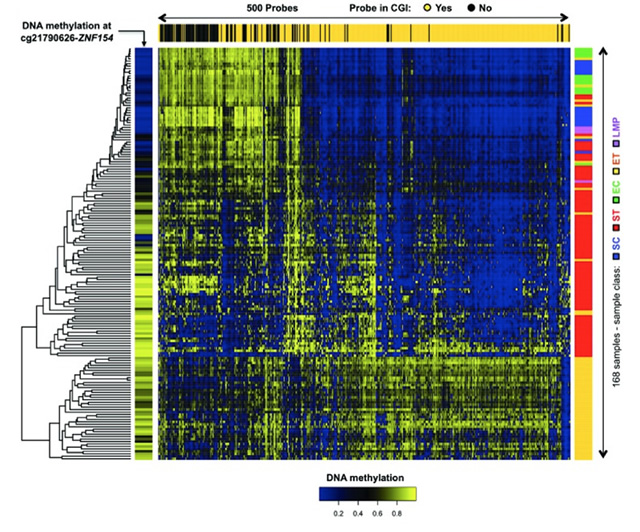
Margolin G, Petrykowska HM, Jameel N, Bell DW, Young AC, Elnitski L. Robust Detection of DNA Hypermethylation of ZNF154 as a Pan-Cancer Locus with in Silico Modeling for Blood-Based Diagnostic Development. J Mol Diagn, (15)00274-3. 2016. [PubMed]
Book Chapters
Yang, M.Q., and Elnitski, L.L. Feature Characterization and Testing of Bidirectional Promoters in the Human Genome - Significance and Applications in Human Genome Research. Chapter 19 in Machine Learning in Bioinformatics, Yan-Qing Zhang and Jagath C. Rajapakse, eds. John Wiley & Sons, 2007.
Yang, M.Q. and Elnitski, L. A computational study of bidirectional promoters in the human genome. Bioinformatics Research and Applications, Zhang, Y., eds, Heidelberg, pp. 361-371. 2007.
Yang, M.Q. and Elnitski, L. Orthology of Bidirectional Promoters Enables Use of a Multiple Class Predictor for Discriminating Functional Elements in the Human Genome. Proceedings of the 2007 International Conference on Bioinformatics & Computational Biology. Arabnia, Yang, M.Q. and Yang, J.Y., eds. pp. 218-228. 2007.
Yang, M.Q., Taylor, J., and Elnitski, L. Rigorous Mapping of Orthologous Bidirectional Promoters in Vertebrates. Proceedings of Computations in Bioinformatics and Bioscience. BMC Bioinformatics Suppl. Issue. pp. 115-122. 2008.
Elnitski L, Burhans R, Riemer C, Hardison R, Miller W. MultiPipMaker: A comparative alignment server for multiple DNA sequences. Current Protocols in Bioinformatics. Wiley & Sons Inc., pp. 10.4.1-10.4.14. 2010.
Welch LR, Koehly LM, Elnitski L. Shared regulatory motifs in promoters of human DNA repair genes. DNA Repair / Book 4, Inna Kruman (Ed.), InTech Publishers. 2011.