Bacterial Artificial Chromosome (BAC)
Definition
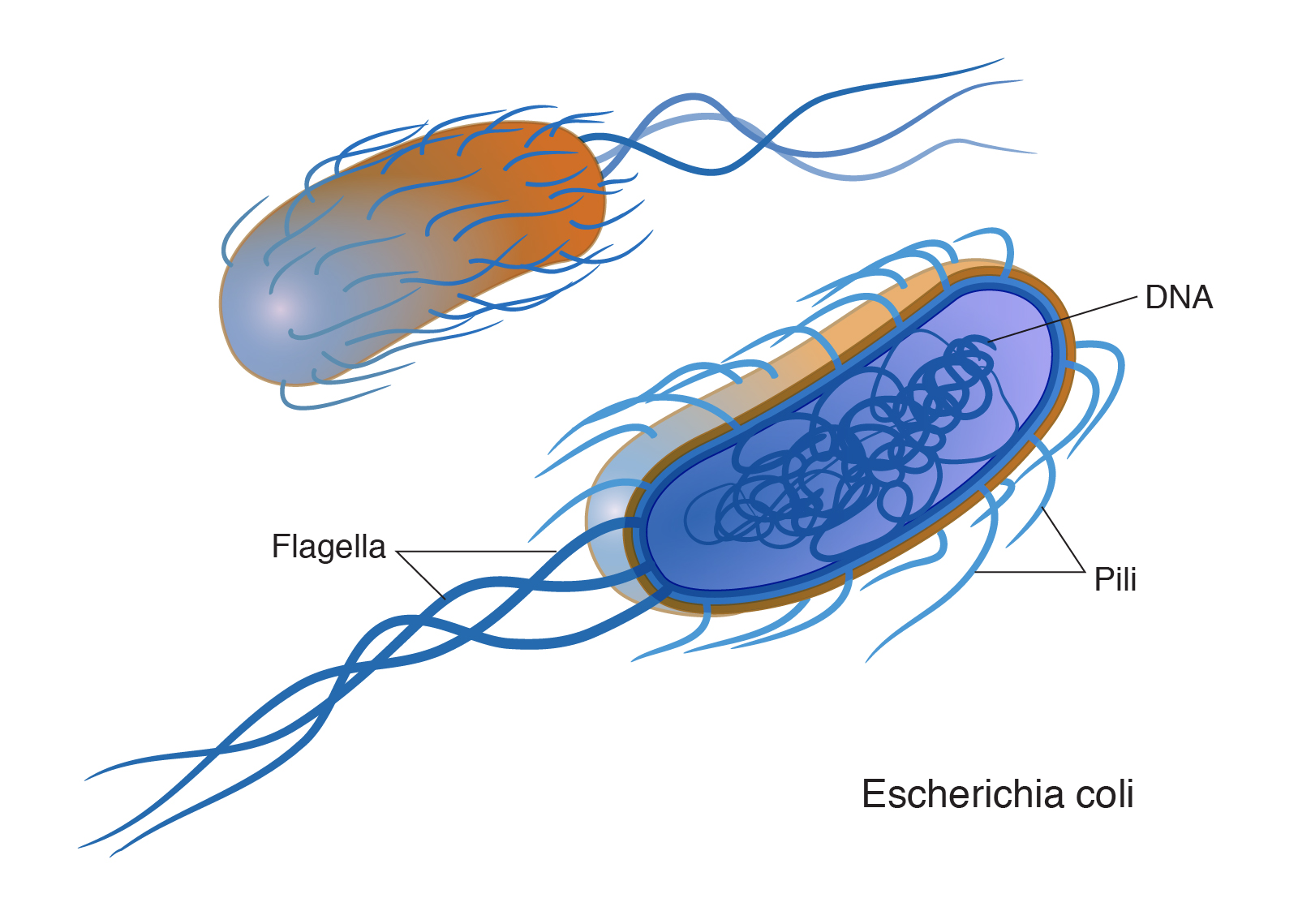
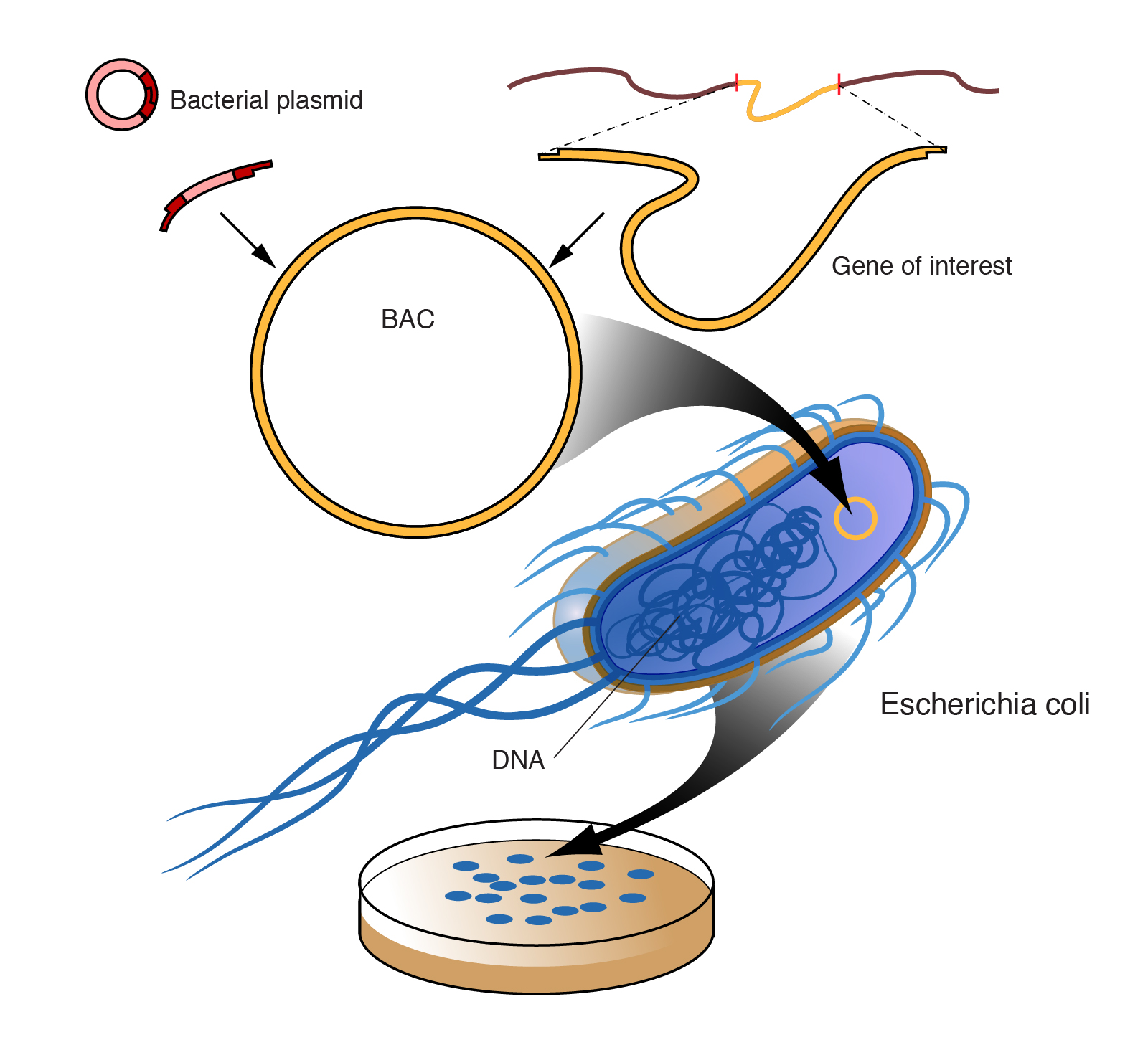
A bacterial artificial chromosome (BAC) is an engineered DNA molecule used to clone DNA sequences in bacterial cells (for example, E. coli). BACs are often used in connection with DNA sequencing. Segments of an organism's DNA, ranging from 100,000 to about 300,000 base pairs, can be inserted into BACs. The BACs, with their inserted DNA, are then taken up by bacterial cells. As the bacterial cells grow and divide, they amplify the BAC DNA, which can then be isolated and used in sequencing DNA.
Narration
A large piece of DNA can be engineered in a fashion that allows it be propagated as a circular artificial chromosome in bacteria--so-called bacterial artificial chromosome, or BAC. Each BAC is a DNA clone containing roughly 100 to 300 thousand base pairs of cloned DNA. Because the BAC is much smaller than the endogenous bacterial chromosome, it is straightforward to purify the BAC DNA away from the rest of the bacteria cell's DNA, and thus have the cloned DNA in a purified form. This and other powerful features of BACs have made them extremely useful for mapping and sequencing mammalian genomes.