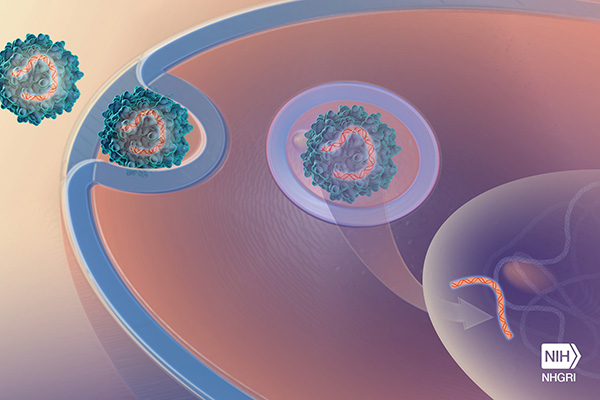
Started in 2013 with support from NHGRI and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the Newborn Sequencing in Genomic Medicine and Public Health (NSIGHT) program has explored implications, challenges, and opportunities associated with the possible use of genome sequence information in the care of newborns. Specifically, NSIGHT researchers have examined the potential benefits of using such information to screen newborn infants for genetic conditions, to further the understanding of diseases that appear during the newborn period, and to improve treatments for these diseases.
Millions of infants born in the U.S. each year are screened for genetic diseases. Traditionally, genome sequencing has not been used for this screening. The recent advances in DNA sequencing technologies have triggered a growing interest in the use of genome sequencing as a newborn screening tool, but more research in this area was needed. Thus, the NSIGHT program was envisioned and launched.
The NSIGHT program funded four pilot projects at seven sites, specifically at Brigham and Women's Hospital, Boston Children’s Hospital, and Baylor College of Medicine; Rady Children’s Hospital and Children's Mercy Hospital; University of California, San Francisco; and University of North Carolina at Chapel Hill. The program’s goals were outlined in a paper published in Pediatrics in February 2017. Overall, the funded NSIGHT projects aimed to address three major research questions:
- Can genome sequencing replicate or augment existing newborn screening methods?
- Would genome sequencing provide insights about clinical conditions not currently screened for in newborns?
- Could additional clinical information be gained from genome sequencing relevant to the clinical care of newborns?
After six years, the NSIGHT program is now coming to an end. To mark this milestone, NSIGHT researchers recently held a final meeting to share their research findings. Overall, NSIGHT found that genome sequencing is a good tool for diagnosing sick infants in the neonatal intensive care unit. In addition, genome sequencing can be complementary to current screening methods for capturing information about some genetic conditions; it may also serve as a good second-tier test for infants that screen positive through current newborn screening procedures.
By design, NSIGHT researchers examined multiple variables in their studies. For example, they sequenced genomes and exomes (the portion of the genome that reflects protein-coding regions), used blood spots and saliva as the source of DNA, enrolled healthy and sick infants into their studies, and focused on different types of conditions (e.g., childhood-onset and medically actionable, adult-onset and medically actionable, and childhood-onset but not medically actionable).
The NSIGHT studies revealed that doctors and families that choose to participate in research found utility in newborn genome sequencing and that returning genetic risk information did not cause undue stress for those opting to receive genomic information. However, genome sequencing failed to detect some disease conditions that were detected by traditional screening methods. Also in one study, compared with genome sequencing, exome sequencing missed diagnosing some known conditions. Another study found that in the neonatal intensive care unit setting, genome sequencing provided a diagnosis for a significant number of babies, some then received treatment, and in at least one case an infant’s life was saved.
Even though the NSIGHT program is coming to an end, some NSIGHT projects are already working to expand their individual studies through collaborations and additional funding. These efforts will continue to examine cost-effectiveness and diagnostic rates for genome sequence as a screening tool; they will also continue to study the implementation of genome sequencing as a rapid method for diagnosing sick infants. If you are interested in more information, a public session was held as part of the final meeting and can be viewed via YouTube.
![]()