Last updated: March 21, 2012
2011 News Features The Road To The 1000 Genome Via Nanopores A Roundup Of Sequencing Technology Developments
The Road to the $1000 Genome Via Nanopores - A Roundup of Sequencing Technology Developments
By Geoff Spencer
NHGRI Staff Writer
February 2011
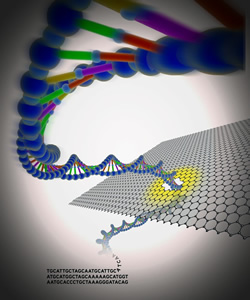 |
Third generation DNA sequencing devices capable of this type of rapid and reliable sequencing have been in development for years. Some of the devices will be able to detect and read DNA molecules as they are driven through nanopores, tiny holes only two nanometers, or about one-billionth of a meter wide, that can distinguish the unique electronic signals of the four bases that make up DNA: Adenine, Thymine, Cytosine and Guanine. The first nanopore-based DNA sequencing devices are predicted to appear by 2014.
Nanopore sequencing, as it is known, is just one of many promising technologies currently being pursued by National Human Genome Research (NHGRI) grantees to achieve high quality human genome sequencing for $1000 or less.
NHGRI launched this effort in 2004 with the award of the first grants from its Advanced DNA Sequencing Technology Program.
At the time, Sanger sequencing could produce a high-quality draft of a human or other mammalian genome using 100 machines for three to four months at a cost of $20 million. The Human Genome Project used Sanger sequencing.
The initial five-year goal of the program was to develop second, or next generation (NextGen) sequencing technologies, that could reduce the cost of sequencing a human genome by two orders of magnitude to $100,000. That sequencing goal was met and surpassed, thanks in part to the NHGRI program and other academic and private efforts. Such efforts have miniaturized the technologies that use less of the expensive chemical reagents needed in the first generation sequencing machines. More importantly, the new technologies are able to simultaneously read many millions, rather than hundreds, of sequencing reactions at once.
NextGen DNA sequencing technologies being used in laboratories today can sequence a human genome on one machine in about a week because these machines process around a billion samples a run. The costs have dropped to about $20,000 per genome.
Despite recent media reports that suggest a $1000 genome is inevitable and could happen as early as this year, difficult hurdles remain. Still, nanopore sequencing shows great promise for dramatically cutting sequencing costs. Among other advantages, it requires little or no chemical reagents or expensive optical systems to detect DNA bases that many current NextGen devices employ.
"For several years, scientists and engineers have been intrigued by the potential to use nanopores as a solution to the low-cost DNA sequencing problem," said Jeffery Schloss, Ph.D., NHGRI's program director for technology development. "Ultimately, we would like these devices to isolate DNA from the tissue or blood sample and rapidly sequence very long DNA molecules. These electronic devices would be simple to use, inexpensive and would connect directly to a computer that would collect and organize the data."
According to Schloss, the promise of obtaining very low cost genomes would allow DNA sequencing to become a routine clinical test, much like blood tests that are conducted as a regular part of healthcare today. And for biomedical researchers, the low cost would allow genome sequences from thousands of people with diseases to be compared to genome sequences from healthy individuals. The ability to sequence genomes at this scale would allow an unprecedented understanding of a variety of diseases such as diabetes, mental health disorders and heart disease.
Researchers are taking varied approaches to overcoming the remaining obstacles for achieving nanopores DNA sequencing. Some researchers are developing methods that use protein or natural nanopores because they are atomically precise. In other words, their diameter is very close to the size of DNA and the inside shape and charge of the nanopores can be manipulated chemically and genetically to perform a novel function - such as recognizing each of the DNA bases - that is different from what they do in nature.
A sequencing method developed by Hagan Bayley, Ph.D., and his colleagues at the University of Oxford, England, relies on a natural protein nanopore derived from a membrane protein of the bacteria, Staphylococcus aureus, called alpha-hemolysin. In the Sept. 8, 2010, issue of Nano Letters, they reported that they modified one of three points inside the nanopore that allowed DNA bases to be accurately identified as they passed through the nanopore.
The following week, in the Proceedings of the National Academy of Sciences, a team led by Michael Niederweis, Ph.D., University of Alabama at Birmingham, and Jens Gundlach, Ph.D., University of Washington, Seattle, distinguished individual nucleotides using a different protein nanopore, the porin A protein from the bacterium, Mycobacterium smegmatis. This group genetically modified the nanopore to detect individual DNA subunits.
In addition to sequencing DNA, nanopore-based devices might be able to perform other analyses on DNA molecules. For instance, the devices might be able to distinguish DNA methylation patterns in the epigenome, chemical markers on DNA that play a central role in normal development as well as diseases such as cancer. Bayley's group reported in the Nov. 21, 2010, issue of the journal Chemical Communications that their protein nanopore, in which a base-discrimination site had been modified, could identify epigenetic DNA modifications.
Protein nanopores are not without problems. "In nature and in the labs developing this technology, protein nanopores sit in a lipid bilayer (the material of which cell membranes are made), which tends to be fragile and easily destroyed," said Schloss. "However, Bayley and Gundlach and others working with protein nanopores are employing elegant solutions to deal with such problems."
Other groups are fabricating artificial or solid-state nanopores that are drilled into insulating material. Several groups are working with atomically thin layers of graphite, called graphene, to craft solid-state nanopores. Graphene is particularly interesting because it might be used as both the nanopore and the electrical connection, making it easier to build sequencing devices. Sheets of graphene are also thin enough to meet the requirement to query a single DNA subunit within a DNA strand.
Jene Golovchenko, Ph.D., Daniel Branton, Ph.D., and their colleagues at Harvard University and the Massachusetts Institute of Technology, reported one of the first demonstrations of transporting DNA through graphene nanopores in the Sept. 9, 2010, issue of Nature. The graphene sheet separates two chambers containing a liquid solution. The team showed that DNA molecules can be detected when they pass through these nanopores, because they block the flow of ions between the liquid chambers.
Another potential strategy to measure DNA bases in nanopores is called electron tunneling. The concept involves a current that flows between two electrodes separated by a small gap. This tunneling current is sensitive to the distance between the electrodes and when a molecule is placed between the electrodes, the current changes and can potentially be used to identify the molecule.
Stuart Lindsay, Ph.D., and colleagues from Arizona State University, have proven that single bases of DNA that are part of a larger strand can be identified using electron tunneling. Their paper, in the Dec. 2010 issue of Nature Nanotechnology, described using chemical 'fingers' that are attached to electrodes and point from either side of a tiny gap, toward the DNA strand passing between them. Those fingers briefly recognize each base and allow an electrical current to pass through, distinguishing among the four DNA bases.
"Since the beginning of the exploration of nanopore sequencing, two modes of detection were envisioned. In one, ions would flow through the pore parallel to the DNA, and different DNA bases would block the ions to a different extent, allowing recognition of each of the base identities," said Schloss. "The second mode is tunneling, in which a current flows across the pore, through the individual bases. This is the first demonstration that tunneling detection works for single nucleotides contained within a DNA strand. Still, there are a number of challenges that must be tackled to build a robust electron tunneling DNA sequencing device."
One issue that must be tackled regardless of which nanopores or measurement scheme is used, is to control the rate of speed at which a DNA molecule moves through a nanopores, called translocation, so it is slowed to the point that single bases are present long enough to be recognized.
Xinsheng Sean Ling, Ph.D. and colleagues at Brown University in Providence, R.I., are developing a solid-state nanopore method that uses short segments of known DNA sequences called probes that match up to corresponding base pairs along the longer DNA molecule meant to be sequenced. Ling's results, published in the Aug. 20, 2010, issue of Nanotechnology, showed that a plastic bead attached to the DNA exerts a drag when the DNA is pulled into the nanopore by an electric field, slowing the DNA's translocation.
Others are exploring the efficacy of placing a DNA polymerase, an enzyme normally involved in DNA replication in cells, at the opening of a nanopore. The polymerase seizes a DNA molecule and acts as a molecular ratchet wrench of sorts, allowing the DNA polymer to advance one nucleotide at a time through the nanopore. With the help of such a tool, each DNA base would remain at a critical location within the nanopore for a length of time measured in milliseconds — enough time for it to be identified.
Mark Akeson, Ph.D., and colleagues at the University of California Santa Cruz, recently demonstrated single base at a time translocation control using three different types of DNA polymerases, in their papers published in Nature Nanotechnology in Nov. 2010 and in the Journal of the American Chemical Society on Dec. 22, 2010. The first paper reports on the precise control of fast electronics used to pull the DNA into the pore, to strike a balance with the activity of the enzyme that was allowing the DNA to move at one DNA subunit at a time. The second paper discusses a different enzyme that permitted use of simpler electronics.
In an article posted in Jan. 2010 in Nano Letters' ASAP page, Bayley's group, designed 'molecular brakes' by genetic engineering a narrow channel of a protein nanopore that slowed DNA translocation speed.
All of the advances by NHGRI-funded investigators and teams and related work in other laboratories are making significant contributions to the development of nanopore DNA sequencing technologies. In the end, Schloss notes, the nanopore platforms and devices that make their way into the clinic would likely make use of a variety of techniques. If these technologies can be incorporated into robust systems, they may some day change life sciences and the practice of medicine, he predicts.
This article is the second in a series of periodic updates that summarizes breakthroughs recently published in the scientific literature by NHGRI-funded researchers who address or overcome obstacles to developing revolutionary third generation sequencing technologies. Read the first article here: The Road to the $1000 Genome — A Roundup of Sequencing Technology Developments.
Last Reviewed: March 21, 2012