Last updated: March 29, 2012
Activity 1: Genetic Variation in Populations
Activity 1: Genetic Variation in Populations
The growing ability to detect and measure human genetic variation allows us to study similarities and differences among individuals. In this activity, you will analyze data on genetic variation and address a series of questions about variation within and between populations.
You should understand the following concepts before you begin this activity:
- the relationship between genes and proteins;
- the relationship between gene and phenotype;
- the difference between a gene and an allele; and
- statements of frequency, for example, 0.45 (45 percent).
Variation in Populations
How do we ordinarily identify a person as a member of a particular racial or ethnic group?
To what extent do we focus on external characteristics such as skin color, hair texture, or characteristic facial structures?
Are there drawbacks to relying on external physical characteristics?
Are there any other biological indicators that could provide insights into group similarities and differences?
Would examining the genetic similarities and differences provide reliable guidance to the classification of groups?
Look at allele frequencies for three different genes in populations around the world. You will see several maps that contain a subset of the actual data collected by scientists.
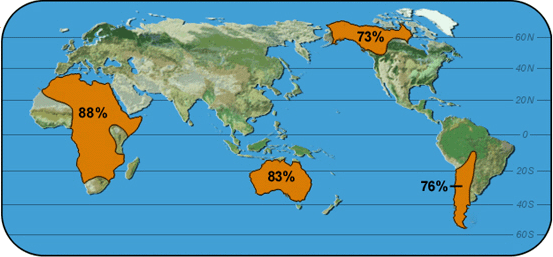 |
Map 1: GC-1 |
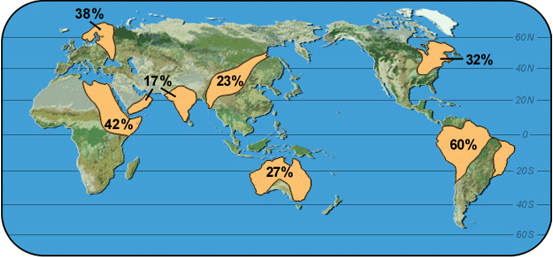 |
Map 1: HP-1 |
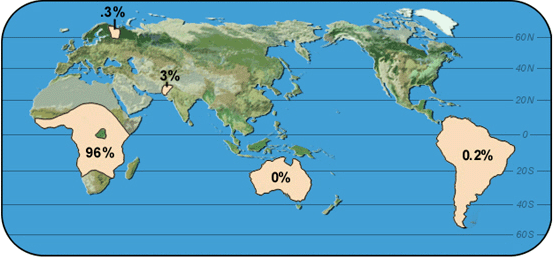 |
Map 1: FY-0 |
What is the range of frequencies for each allele shown?
Which allele varies most in frequency?
Which allele varies least?
Propose some hypotheses that explain the variation in the frequency of FY-0.
Hint 1: Think of hypotheses based on evolution and natural selection.
Hint 2: Natural selection is a function of environmental variations acting on naturally occurring genetic variations and their phenotypic effects.
See the following map to examine additional data to help you refine your hypothesis.
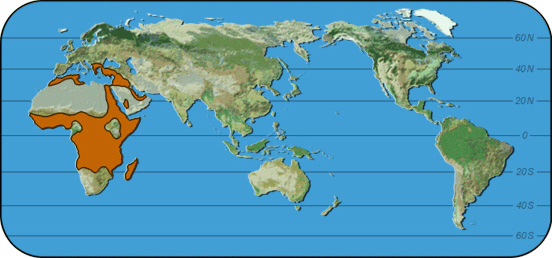 |
Map 2: High incedence of Plasmodium vivax malaria worldwide |
What is the relationship between the incidence of Plasmodium vivax malaria and the FY-O allele?
Review the information below about what scientists know about each of the alleles used in this activity. If you wish, rework your hypothesis.
The GC gene (which has two major alleles, 1 and 2) codes for a blood protein that attaches to vitamin D and regulates its distribution within the body.
The HP gene codes for another blood protein (haptoglobin). This protein attaches to the hemoglobin released by the red blood cells when they decay at the end of their natural life or when they are destroyed by a disease, such as malaria.
The FY gene codes for a protein that is normally found on the surface of red blood cells. The protein makes it easier for a particular malarial parasite, Plasmodium vivax, to get into the red blood cell. Once in the red blood cell, P. vivax, like all malarial parasites, multiplies. The FY-O allele results in the absence of the protein. That makes it hard for the parasite to gain entry to the red blood cells and multiply. Therefore, the FY-O allele provides a certain amount of protection against this type of malaria.
The FY-0 allele provides a selective advantage in regions where Plasmodium vivax malaria is common. That advantage accounts for the increased frequency of the FY-0 allele in those regions. We know of only a few such examples where there is a clear relationship between an environmental variable and differences in allele frequency.
Scientists have examined DNA from chromosomal regions that have a lot of detectable differences in sequence.
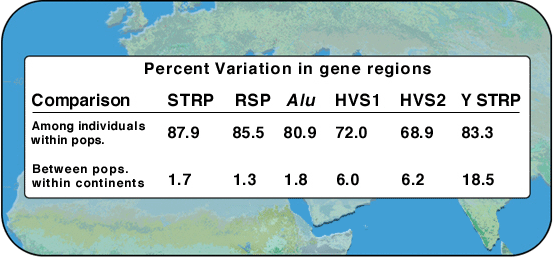 |
Table 1: Percentage of variation at two different levels of population structure |
Definitions:
STRP: autosomal short-tandem-repeat polymorphisms. These are variable segments of DNA that are 3, 4 or 5 bases long, repeated over and over.
RSP: autosomal restriction-site polymorphism. DNA sequence variations that occur at a restriction enzyme recognition sequences that result in variations (polymorphisms) in the length of DNA fragments obtained by cutting DNA with a particular restriction enzyme.
Alu: Alu-insertion polymorphism. Alu sequences are repeat sequences that are about 300 bases long. There are many thousands of Alu repeats in the genome, and they appear within genes and between genes. They have no known function.
HVS1 and HVS2: hypervariable sequence 1 and 2. This is DNA from mitochondria taken from regions that have a lot of differences in DNA sequence.
Y-STRP: Y chromosome short-tandem-repeat polymorphisms.
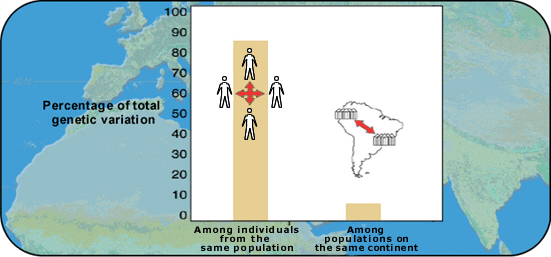 |
Figure 1: Percentage of variation at two different levels of population structure |
Now examine the data above from a study of worldwide genetic variation in 255 individuals. The individuals included 72 Africans, 63 Asians and 120 Europeans.
Based on the data you have seen, could you draw boundaries that would separate populations clearly on the basis of genetic differences?
The study of genetic variations in Homo sapiens shows that there is more genetic variation within populations than between populations. This means that two random individuals from any one group are almost as different as any two random individuals from the entire world.
Although it may be easy to observe distinct external differences between groups of people, it is more difficult to distinguish such groups genetically, since most genetic variation is found within all groups.
Footnotes
1. Allele: an alternate form of a gene at a given locus. For example, the gene for ABO blood group has three alleles, A, B, and O.
2. Allele frequency: the commonness of a particular allele in a given population, stated as a number, from 0 to 1, or as a percentage, from 0 to 100.
