Last updated: February 28, 2012
NHGRI Research Reveals Humans Share a Common Set of Skin Microbes
NHGRI Research Reveals Humans Share a Common Set of Skin Microbes
Study Also Finds Similarity Between Microbes of Human and Mouse Skin
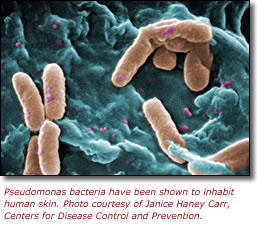 Our skin is crawling with millions of tiny microorganisms that play a critical role in skin health and disease. A team of researchers from the National Institutes of Health (NIH), led by Julie Segre, Ph.D., a senior investigator in the Genetics and Molecular Biology Branch at the National Human Genome Research Institute (NHGRI), has published a study in the May 23 online issue of Genome Research. The researchers found that healthy humans share a common set of microorganisms living in and on the area of skin around the inner elbow, an area where atopic dermatitis (eczema) is known to develop.
Our skin is crawling with millions of tiny microorganisms that play a critical role in skin health and disease. A team of researchers from the National Institutes of Health (NIH), led by Julie Segre, Ph.D., a senior investigator in the Genetics and Molecular Biology Branch at the National Human Genome Research Institute (NHGRI), has published a study in the May 23 online issue of Genome Research. The researchers found that healthy humans share a common set of microorganisms living in and on the area of skin around the inner elbow, an area where atopic dermatitis (eczema) is known to develop.
The human body hosts a diverse set of microbial communities at different body sites. Not very much is known about the fundamental role microorganisms play in human health and disease. Moreover, microbiologists cannot isolate and grow many of these different micro-organisms, making them difficult to study. So new techniques have been developed in which researchers use genetic sequencing to analyze microbial genomes, collectively called a microbiome, to determine what is actually living in and on our skin.
NIH recently launched the Human Microbiome Project, a part of the NIH Roadmap for Medical Research [commonfund.nih.gov/], to discover what microbial communities exist in different parts of the human body, including the skin, and explore how these communities change in the presence of health or disease. Understanding the role of microbes in human health and disease could transform how patients are diagnosed and treated in the future.
Dr. Segre, whose laboratory's research focuses on the skin and diseases such as eczema and psoriasis, said: "We specifically looked at the inner elbow to lay the groundwork for future studies elucidating the genetic and environmental factors of the extremely common inflammatory skin disorder atopic dermatitis, which affects this area of the skin and is associated with Staphylococcus infections."
In addition to Dr. Segre and other NHGRI researchers, the paper includes authors from the National Cancer Institute and the NIH Intramural Sequencing Center. The work was supported by NHGRI, NCI, and the National Institute of General Medical Sciences, all part of NIH.
In order to characterize the microbiome of the inner elbow, the researchers collected samples from the left and right inner elbows of five healthy human volunteers using three different methods (swab, scrape, and punch biopsy) to survey the microbiome at different levels of the skin. Using genetic technologies which can distinguish and identify microbes, the researchers found that the majority of inner elbow microbes were Proteobacteria, primarily Psuedomonas and Janthinobacterium. Both microbes belong to the family of Psuedomonad bacteria, a somewhat surprising finding because they are not traditionally thought of as skin microbes. However, these bacteria are widespread in nature and flourish in moist and sometimes sweaty environments, so researchers were not surprised that they reside on the inner elbow of humans.
The research also showed that a core skin microbiome exists among the five individuals studied, comprising the majority of microbes from six bacterial divisions, including the Pseudomonad's referred to above, which are the two most common bacterial divisions. "This finding is critical for the future study of disease," Dr. Segre explained. "Our long-term goal is to study how dynamic changes in the skin microbiome are correlated to skin disorders associated with known and suspected bacterial infections."
Interestingly, the researchers then collected samples from the ear skin of a laboratory mouse because this site shares a lot in common with most regions of the human skin. Researchers report this particular line of mice, used in studies of skin inflammation, could potentially be used as a model for understanding the interactions of the genetic and environmental contribution to skin diseases, which could potentially lead to novel drugs and treatments.
Based on their findings, the research team has recently launched another study to sample 20 skin sub sites in healthy individuals in order to lay the groundwork for explaining why various skin diseases occur at specific sites, such as psoriasis on the outside of and eczema on the inside of elbows and knees. The researchers hope any results will be used to inform which skin sites are selected as part of the Human Microbiome Project and also plan to make their data available to the Human Microbiome Project as part of its collection of 1,000 microbial genomes collection.
Last Updated: February 28, 2012
