Last updated: March 15, 2012
Using DNA Sequencing to Monitor Organ Transplant Rejection
Genome Advance of the Month
Using DNA Sequencing to Monitor Organ Transplant Rejection
April 2011
By Jonathan Gitlin, Ph.D.
Science Policy Analyst
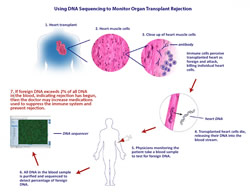
Thomas Snyder and his colleagues demonstrated that it is possible to identify organ rejection noninvasively by detecting DNA from the transplanted organ — which is essentially foreign genetic material — released into the recipient's bloodstream when the patient's immune system attacks the transplanted organ as if it were a dangerous infection. This is yet another example of how the genome-analyzing technologies developed during Human Genome Project, and ever-decreasing costs for DNA sequencing, are beginning to impact the field of medicine.
The history of organ transplantation goes back a long way, but until the development of immunosuppressive drugs, allotransplantation (transplanting tissue from a donor with different DNA) always ended in failure. Basically, the recipient's immune system recognizes the donor tissue as foreign and mounts a massive attack to destroy the invader (the transplanted organ), resulting in rejection of the organ. Understanding the mechanism behind rejection and developing drugs such as cyclosporin to suppress it, enabled surgeons to transplant organs with much greater success. Even though transplantation patients now take these powerful immunosuppressants, the drugs do not always prevent tissue rejection.
In the case of transplanted hearts, doctors determine the health of the new organ through a procedure called endomyocardial biopsy. Basically, a tube is threaded through blood vessels until it reaches the heart and a small piece of heart tissue is cut out and retrieved for analysis. When the immune system starts to attack a transplant organ, it starts slowly, killing some of the individual cells in the transplanted heart, which can be seen in the biopsy sample.
Scientists have been working on other, noninvasive methods, mainly by attempting to gauge the activity of the donor's immune response. The novelty in Snyder et al.'s approach is that the Stanford team detects the presence of donor DNA in the recipient's blood as a marker for organ damage. During rejection, cells in the transplanted organ begin to die in a process called apoptosis, releasing fragments of foreign DNA into the blood.
In cases where female patients (who have two X chromosomes) have received organs from male donors (who have one X and one Y chromosome), it has been possible to detect the presence of DNA from the Y chromosome, but female patients who have received male organs make up less than a quarter of transplant patients, so a universal method represents a significant step forward. In this study, blood samples were obtained from female heart transplant patients, the DNA was purified and then sequenced.
Donor DNA was identified by looking for single nucleotide polymorphisms that the patient was homozygous for but that the donor was either heterozygous, or homozygous with a different variant. For example, if both of the recipient's genes had an A at the site in question, but the donor had one copy with an A and one with a T, or two copies with T, finding a T during the sequencing would indicate the presence of donor DNA.
When the patients were healthy and rejection was under control, less than 0.5 percent of the DNA detected in the blood was from the donor heart. The study found that above a threshold of two percent donor DNA in the blood, there was an 80 percent true positive rate for tissue rejection, along with a 15 percent false positive rate. They were also able to demonstrate an increase in the amount of donor DNA detected over time before the onset of rejection symptoms, which other techniques under consideration have not been able to accomplish. Because donor DNA can be detected earlier than other rejections signals, physicians might use that information to increase immune suppression to get ahead of a rejection event before it happens, keeping both the transplanted organ and the patient recipient healthier.
"I am excited to see unexpected clinical applications are emerging from the Human Genome Project and the development of next generation sequencing technologies," said Dr. Stephen Quake, senior author on the paper. While this technique isn't ready to be rolled out into the clinic just yet, it represents a promising new tool to help in the treatment of organ transplant patients.
Read the study: Universal noninvasive detection of solid organ transplant rejection, Proc. Nat. Acad. Sci. U.S.A., April 12, 2011
April also saw the first publication from an exciting NHGRI program that is integrating electronic medical records and DNA biobanks. The Electronic Medical Records and Genomics (eMERGE) Network is a multi-center initiative that involves the Group Health Cooperative of the University of Washington, Marshfield Clinic, Mayo Clinic, Northwestern University, and Vanderbilt University. Each institution is linking patients' electronic medical records to matched DNA samples in order to use those resources to explore the relationship between genetic variation and a particular disease or physical trait; dementia, cataracts, diabetes, peripheral artery disease, and cardiac conductance.
The paper, published in Science Translational Medicine, showed that it is possible to identify patients with the desired phenotype with good enough accuracy for use in genome-wide association studies. Although this is the first time it has been possible to do this, with optimization and a wider use of electronic medical records, it is an approach that could have huge ramifications for clinical decision support and personalized medicine.
Read the study: Electronic Medical Records for Genetic Research: Results of the eMERGE Consortium [ncbi.nlm.nih.gov], Sci. Trans. Med., April 20, 2011