Last updated: August 27, 2013
Fitting the National Archives in your pocket
Genome Advance of the Month
Fitting the National Archives in your pocket
By Joy Yang
Post-baccalaureate Fellow
 |
Nick Goldman, Ph.D., and Ewan Birney, Ph.D., researchers at the European Bioinformatics Institute in Hinxton, UK, wandered off on a tangent after a couple of beers. Instead of their usual mission of decoding genomic information, they decided to try the opposite problem: encoding information in DNA.
"This is one of the best moments in science - it starts off with an idea over a beer, and ends up as a letter to Nature," Dr. Birney said.
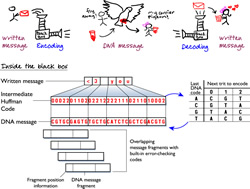
Intuitively, because there are four bases (or "letters") that make up the alphabet of DNA, each base in a DNA sequence can be used to represent one of four possibilities. For example, to represent a binary code, A (Adenine) could represent 00, C (Cytosine) could represent 01, G (Guanine) could be 10 or T (Thymine) could be 11. However, this strategy will not get you the 0 percent error rate that Drs. Goldman and Birney wanted to achieve.
That's because some DNA sequences are more difficult to sequence than others. When a DNA sequence contains homopolymers -or long runs of the same base, like TTTTT - it can be difficult to tell exactly how many bases are contained in that stretch. So, the researchers devised a clever encoding scheme to prevent any repetition of the same base.
In addition, because long messages are more likely to contain errors than short messages, they broke each message into fragments with codes containing information about the fragment, such as what position the fragment takes in the entire message, how long the information encoded in the fragment is and a few other error-checking codes. This helps the researchers filter out fragments of information containing errors from either synthesis (writing information to DNA) or sequencing (reading information from DNA).
A more complete description of their strategy is depicted in the cartoon above.
Using this method, Drs. Goldman and Birney synthesized DNA versions of all 154 of Shakespeare's sonnets, a PDF of Watson and Crick's paper on the structure of DNA, a photograph of the European Bioinformatics Institute, an mp3 of Martin Luther King's "I Have a Dream" speech and the key for the Huffman Code. The DNA samples were then decoded with 100 percent accuracy. The researchers did, however, need to take special means to repair the PDF document, where the DNA code contained long, repeating sequences that folded on itself, making it difficult to sequence. However, the researchers already have ideas that will eliminate this issue.
While this study is the most successful approach at encoding messages in DNA, this problem has previously been tackled for various intriguing applications. For example, in 1999 researchers at Mount Sinai School of Medicine in New York City showed that DNA can be used as a tool for espionage. They demonstrated this by concealing a short message, "June 6 invasion: Normandy," in human genomic DNA, and placing the DNA on a period at the end of a sentence in a written letter.
Also in 1999, and commissioned by Ars Electronica as art, Eduardo Kac encoded a sentence from the biblical book of Genesis in a plasmid, a small DNA molecule that can replicate independently of bacterial DNA and exchanged between bacteria. It also contained the sequence for a fluorescent protein. This plasmid was inserted into the bacteria E. coli and the fluorescence was used to trace the message as it was duplicated, or passed, from one bacteria to another.
Just last August, George Church, Ph.D., professor of genetics at Harvard Medical School, encoded his new book, Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves, using DNA microchips. According to Science Daily, Dr. Church had considered including DNA copies of the book in each print copy but decided against it due to unexplored issues in safety. For example, while short DNA fragments are unlikely to be harmful on their own, they can be absorbed by microbes in the environment, and these results can be unpredictable. In addition, seemingly harmless computer codes may represent human viruses, or seemingly harmless DNA code may represent computer viruses. In fact, the researchers were able to encode a spyware program in DNA to demonstrate this possibility.
Returning to our Genome Advance of the Month, Drs. Goldman and Birney calculate that using this method, the entire collection of the U.S. National Archives (100 terabytes of data) could be encoded in 0.05 g of DNA, which is less than 1/50 the mass of a penny. That doesn't mean, however, that bookstores will be selling entire libraries in test tubes just yet. While DNA is stable and requires very little maintenance (after all, we have been able to sequence the Woolly Mammoth genome), writing information to, and reading information from DNA is still much more expensive than writing, reading and maintaining magnetic tape. So, Drs. Goldman and Birney estimate that it is currently more cost effective to store data in DNA only if the data will not be accessed for 600 to 5,000 years. However, if the cost of synthesizing DNA continues to decline as it has, it may not be too long before this number drops to 50 years.
Drs. Goldman and Birney envision their study to be useful, mainly for archival purposes. And while the cost of synthesizing DNA remains high, the two are already plotting to place the world on a "string" (a term used in computer programming to mean a sequence of symbols from the same alphabet, which in this case would be DNA created by an alphabet consisting of the four bases A, C, G and T) by creating a DNA time capsule for archiving the world's data.
All they need now is an altruistic billionaire to fund the project.
Read the articles
- Goldman, N., et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature, 2013. [Full Text]
- Clelland, C.T., V. Risca, and C. Bancroft. Hiding messages in DNA microdots. Nature, 399(6736): p. 533-534. 1999. [Full Text]
- Kac, E. Genesis. Ars Electronica 99. 1999. [cited 2013 Feb 19]; Available from: [Genesis]
- Church, G.M., Y. Gao, and S. Kosuri. Next-Generation Digital Information Storage in DNA. Science, 2012. 337(6102): p. 1628. [PubMed]
