
Alexander Wilson, Ph.D.
Division of Intramural Research
B.A. Western Maryland College
Ph.D. Indiana University
Biography
Dr. Alexander F. Wilson graduated from McDaniel College, magna cum laude, with a B.A. in biology in 1975, and received the H.P. Studivant Award as the Outstanding Biology Major. He received his Ph.D. in medical genetics from Indiana University under the direction of P. Michael Conneally, Ph.D., (1980) and did his postdoctoral training in statistical genetics with Robert C. Elston, Ph.D., in the Department of Biometry, Louisiana State University Medical Center (1980-1982). He remained at Louisiana State University, rising to the rank of tenured Full Professor in 1993. He was recruited to the National Human Genome Research Institute in 1995.
He is a scientist emeritus in the Computational and Statistical Genomics Branch, at the National Human Genome Research Institute, NIH, and an adjunct professor, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health. Dr. Wilson is an active member of the American Society of Human Genetics (ASHG) and the International Genetic Epidemiology Society (IGES). He has served on the ASHG Program Committee (2010-2013), on the IGES Board of Directors, and is currently the President of IGES.
He has been a member of or has directed the dissertation committees of 16 students and has trained over a dozen post-doctoral students and visiting faculty. He has received numerous awards, including the Indiana University School of Medicine Department of Medical Genetics Distinguished Alumnus Award, the Western Maryland College Trustee Alumni Award, the NIH Director's Award and induction into Phi Beta Kappa as an alumni member.
His research interests focus on the identification of genetic effects that may be responsible for phenotypic variation in quantitative traits (e.g., traits related to cardiovascular disease and scoliosis), the coding and non-coding elements that may be responsible for their expression, and the investigation of the statistical properties of newly developed methods of genetic analysis for quantitative traits.
-
Biography
Dr. Alexander F. Wilson graduated from McDaniel College, magna cum laude, with a B.A. in biology in 1975, and received the H.P. Studivant Award as the Outstanding Biology Major. He received his Ph.D. in medical genetics from Indiana University under the direction of P. Michael Conneally, Ph.D., (1980) and did his postdoctoral training in statistical genetics with Robert C. Elston, Ph.D., in the Department of Biometry, Louisiana State University Medical Center (1980-1982). He remained at Louisiana State University, rising to the rank of tenured Full Professor in 1993. He was recruited to the National Human Genome Research Institute in 1995.
He is a scientist emeritus in the Computational and Statistical Genomics Branch, at the National Human Genome Research Institute, NIH, and an adjunct professor, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health. Dr. Wilson is an active member of the American Society of Human Genetics (ASHG) and the International Genetic Epidemiology Society (IGES). He has served on the ASHG Program Committee (2010-2013), on the IGES Board of Directors, and is currently the President of IGES.
He has been a member of or has directed the dissertation committees of 16 students and has trained over a dozen post-doctoral students and visiting faculty. He has received numerous awards, including the Indiana University School of Medicine Department of Medical Genetics Distinguished Alumnus Award, the Western Maryland College Trustee Alumni Award, the NIH Director's Award and induction into Phi Beta Kappa as an alumni member.
His research interests focus on the identification of genetic effects that may be responsible for phenotypic variation in quantitative traits (e.g., traits related to cardiovascular disease and scoliosis), the coding and non-coding elements that may be responsible for their expression, and the investigation of the statistical properties of newly developed methods of genetic analysis for quantitative traits.
Scientific Summary
The overarching goal of Dr. Wilson's research program has been the identification of genetic variants responsible for the variation in quantitative traits. The specific aims of this research program are: 1) to use statistical genetic analysis to identify genetic effects underlying quantitative traits and to identify statistical challenges that need to be addressed, 2) to develop new methods of genetic analysis for quantitative traits that address these challenges, 3) to use computer simulation to investigate the statistical properties of these methods, and 4) to apply insights gained from these simulations to ongoing collaborative studies. This work has spanned more than three decades, first at the Department of Biometry and Genetics at the Louisiana State University Medical Center and then the Genometrics Section of the Computational and Statistical Genomics Branch of the NHGRI Division of Intramural Research, NIH. This research has included applications projects and methods development in linkage analysis and tests of association with red-cell antigens, protein polymorphisms, STRPs, SNPs and next-generation sequence variants (SVs), in both family- and population-based samples.
Major substantive results include 1) the identification of genes for Mendelian syndromes (e.g., congenital cataracts, Cranio-Lenticulo-Sutural-Dysplasia), 2) the identification of polymorphisms responsible for variation in quantitative traits (e.g., dopamine-beta-hydroxylase activity, citalopram response in depressed individuals, and platelet aggregation), and 3) the identification of candidate regions with linkage and association in complex disorders (e.g., traits related to hypertension and cardiovascular disease, depression and alcoholism, familial idiopathic scoliosis and kyphoscoliosis, and craniosynostosis).
Methodological work includes: 1) advances in non-parametric linkage analysis, 2) stepwise regression of identified variants in quantitative results, 3) regional inference with moving averages of p-values, 4) the use of derived composite biallelic loci, 5) testing associations in parent-offspring trios with a regression of offspring on mid-parent (ROMP) based approach, and 6) the use of hot-spot based delimiters to divide the genome into independent segments in a linear regression format for family- and population-based tests of association for sequence variants (tiled regression). In addition, the large and small sample statistical properties of these tests have been investigated with computer simulation studies to ensure that the tests are statistically valid and have reasonable power and type I error rates. Software packages developed include 1) the Genometric Simulation Analysis Package (GASP), 2) the implementation of the Regression of Offspring on Mid-Parent (ROMP, ROOP and ROMPrev) and 3) the implementation of the tiled regression approach, the Tiled Regression Analysis Package (TRAP). These packages are available on the NHGRI website.
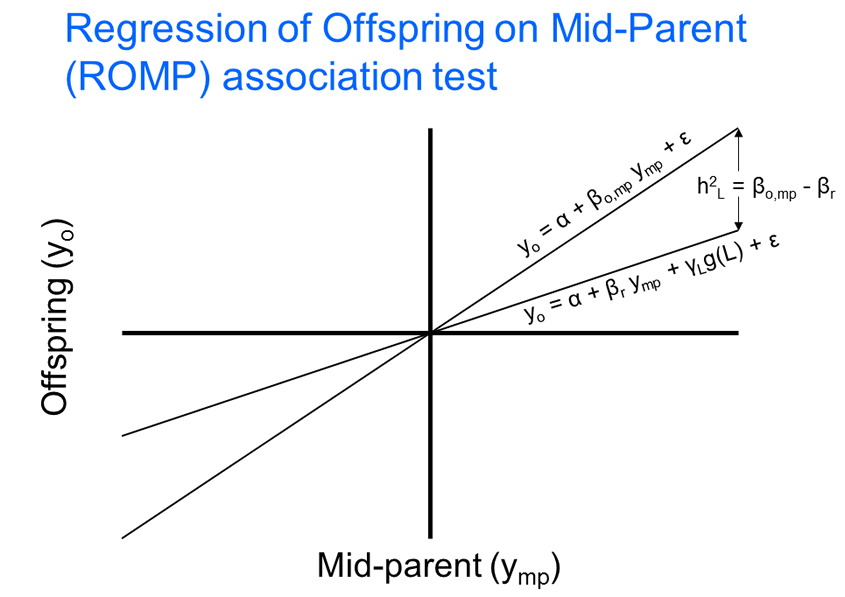
During the last several years, the density of genetic variants has increased dramatically and the section's research has become particularly focused on approaches that are robust with respect to the non-independence between markers and correlations between family members within samples, and on the identification of non-coding regulatory elements. Recent methodological work has focused on the development of two new methods for quantitative traits: the regression of offspring on mid-parent (ROMP), and tiled regression. Both are tests of association in a linear regression framework that can be applied to family data and have been designed for data with very large numbers of genetic markers (millions), e.g. high density SNP panels, and/or large-scale sequencing producing large numbers of rare sequence variants. ROMP is designed to minimize the amount of genotyping and/or sequencing required for a test of association in a parent-offspring trio or nuclear family, by requiring phenotyping data on the parents and offspring, but requiring genotyping or sequencing on only the offspring. Tiled regression is a method that determines the set of independent sequence variants across the entire genome that best predict a given phenotype, against the background of all the variants in the genome.
Regression methods are used to identify independent variants (both coding and non-coding) in predefined independent tiles, that are defined by hotspot blocks, or other positional or functional regions. Higher level regression is then used to determine independent variants over chromosomes and the entire genome. At the Genetic Analysis Workshop 17 (GAW 17), in 2010, it was discovered that there was a substantial inflation of type I error when GWAS methods were used to analyze rare sequence variants, most likely due to the presence of gametic disequilibrium (or inter-LD). The tiled regression method was one of the only methods that allowed for both intra- and inter-LD correlations and it did not exhibit inflation of type I error rates that were present in virtually all of the other methods considered. Before this workshop, it was generally assumed that adjustments only had to be made for correlations within LD blocks (intra-LD correlations); but based on the GAW 17 findings, this is clearly not the case.
Future plans include extensions of TRAP to qualitative traits, and the inclusion of tiled regression into the regression of offspring on mid-parent (ROMP) regression framework. The tiled regression approach, using both hotspot and functional criteria to define tiles, is currently being used in all of our ongoing and future collaborative studies. These projects include 1) analysis of all ClinSeq traits and sequence data (Les Biesecker et al.), 2) analysis of the Trinity Irish metabolite data (Lawrence Brody), 3) functional studies in zebrafish based on our kyphoscoliosis IRX results (Nancy Miller), 4) whole exome sequencing and analysis of a large family with metopic craniosynostosis (Simeon Boyadjiev), and 5) targeted or whole exome sequencing and analysis of about 70 families with familial idiopathic scoliosis (Nancy Miller).
-
Scientific Summary
The overarching goal of Dr. Wilson's research program has been the identification of genetic variants responsible for the variation in quantitative traits. The specific aims of this research program are: 1) to use statistical genetic analysis to identify genetic effects underlying quantitative traits and to identify statistical challenges that need to be addressed, 2) to develop new methods of genetic analysis for quantitative traits that address these challenges, 3) to use computer simulation to investigate the statistical properties of these methods, and 4) to apply insights gained from these simulations to ongoing collaborative studies. This work has spanned more than three decades, first at the Department of Biometry and Genetics at the Louisiana State University Medical Center and then the Genometrics Section of the Computational and Statistical Genomics Branch of the NHGRI Division of Intramural Research, NIH. This research has included applications projects and methods development in linkage analysis and tests of association with red-cell antigens, protein polymorphisms, STRPs, SNPs and next-generation sequence variants (SVs), in both family- and population-based samples.
Major substantive results include 1) the identification of genes for Mendelian syndromes (e.g., congenital cataracts, Cranio-Lenticulo-Sutural-Dysplasia), 2) the identification of polymorphisms responsible for variation in quantitative traits (e.g., dopamine-beta-hydroxylase activity, citalopram response in depressed individuals, and platelet aggregation), and 3) the identification of candidate regions with linkage and association in complex disorders (e.g., traits related to hypertension and cardiovascular disease, depression and alcoholism, familial idiopathic scoliosis and kyphoscoliosis, and craniosynostosis).
Methodological work includes: 1) advances in non-parametric linkage analysis, 2) stepwise regression of identified variants in quantitative results, 3) regional inference with moving averages of p-values, 4) the use of derived composite biallelic loci, 5) testing associations in parent-offspring trios with a regression of offspring on mid-parent (ROMP) based approach, and 6) the use of hot-spot based delimiters to divide the genome into independent segments in a linear regression format for family- and population-based tests of association for sequence variants (tiled regression). In addition, the large and small sample statistical properties of these tests have been investigated with computer simulation studies to ensure that the tests are statistically valid and have reasonable power and type I error rates. Software packages developed include 1) the Genometric Simulation Analysis Package (GASP), 2) the implementation of the Regression of Offspring on Mid-Parent (ROMP, ROOP and ROMPrev) and 3) the implementation of the tiled regression approach, the Tiled Regression Analysis Package (TRAP). These packages are available on the NHGRI website.

During the last several years, the density of genetic variants has increased dramatically and the section's research has become particularly focused on approaches that are robust with respect to the non-independence between markers and correlations between family members within samples, and on the identification of non-coding regulatory elements. Recent methodological work has focused on the development of two new methods for quantitative traits: the regression of offspring on mid-parent (ROMP), and tiled regression. Both are tests of association in a linear regression framework that can be applied to family data and have been designed for data with very large numbers of genetic markers (millions), e.g. high density SNP panels, and/or large-scale sequencing producing large numbers of rare sequence variants. ROMP is designed to minimize the amount of genotyping and/or sequencing required for a test of association in a parent-offspring trio or nuclear family, by requiring phenotyping data on the parents and offspring, but requiring genotyping or sequencing on only the offspring. Tiled regression is a method that determines the set of independent sequence variants across the entire genome that best predict a given phenotype, against the background of all the variants in the genome.
Regression methods are used to identify independent variants (both coding and non-coding) in predefined independent tiles, that are defined by hotspot blocks, or other positional or functional regions. Higher level regression is then used to determine independent variants over chromosomes and the entire genome. At the Genetic Analysis Workshop 17 (GAW 17), in 2010, it was discovered that there was a substantial inflation of type I error when GWAS methods were used to analyze rare sequence variants, most likely due to the presence of gametic disequilibrium (or inter-LD). The tiled regression method was one of the only methods that allowed for both intra- and inter-LD correlations and it did not exhibit inflation of type I error rates that were present in virtually all of the other methods considered. Before this workshop, it was generally assumed that adjustments only had to be made for correlations within LD blocks (intra-LD correlations); but based on the GAW 17 findings, this is clearly not the case.
Future plans include extensions of TRAP to qualitative traits, and the inclusion of tiled regression into the regression of offspring on mid-parent (ROMP) regression framework. The tiled regression approach, using both hotspot and functional criteria to define tiles, is currently being used in all of our ongoing and future collaborative studies. These projects include 1) analysis of all ClinSeq traits and sequence data (Les Biesecker et al.), 2) analysis of the Trinity Irish metabolite data (Lawrence Brody), 3) functional studies in zebrafish based on our kyphoscoliosis IRX results (Nancy Miller), 4) whole exome sequencing and analysis of a large family with metopic craniosynostosis (Simeon Boyadjiev), and 5) targeted or whole exome sequencing and analysis of about 70 families with familial idiopathic scoliosis (Nancy Miller).
Publications
Wilson AF, Cohen JC. Hypotheses for testing deviations from random integration: evidence for nonrandom retroviral integration. Genomics, 3(2):137-142. 2008. [PubMed]
Amos CI, Elston RC, Wilson AF, Bailey-Wilson JE. A more powerful robust sib-pair test of linkage for quantitative traits. Genet Epidemiol, 6(3):435-449. 1989. [PubMed]
Wilson AF, Elston RC, Sellers TA, Bailey-Wilson JE, Gersting JM, Deen DK, Sorant AJM, Tran LD, Amos CI, Siervogel RM. Stepwise oligogenic segregation and linkage analysis illustrated with dopamine-beta-hydroxylase activity. Am J Med Genet, 35:425-432. 1990. [PubMed]
Wilson AF, Elston RC, Tran LD, Siervogel RM. Use of the robust sib-pair method to screen for single-locus, multiple-locus, and pleiotropic effects: application to traits related to hypertension. Am J Hum Genet, 48:862-872. 1991. [PubMed]
Wilson AF, Elston RC. Statistical validity of the Haseman-Elston sib-pair test in small samples. Genet Epidemiol, 10(6):593-598. 1993. [PubMed]
Goldin LR, Chase GA, Wilson AF. Regional inference with averaged P values increases the power to detect linkage. Genet Epidemiol, 17(3):157-164. 1999. [PubMed]
Wilson AF, Sorant AJ. Equivalence of single- and multilocus markers: power to detect linkage with composite markers derived from biallelic loci. Am J Hum Genet, 66(5):1610-1615. 2000. [PubMed]
Miller NH, Justice CM, Marosy B, Doheny KF, Pugh E, Zhang J, Wilson AF. Identification of candidate regions in familial idiopathic scoliosis. Spine 30(10):1181-1187. 2005. [PubMed]
McMahon FJ, Buervenich S, Charney D, Lipsky R, Rush AJ, Wilson AF, Sorant AJM, Papanicolaou GJ, Laje G, Fava M, Trivedi M, Wisniewski S, Manji H. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of citalopram treatment: Results from the STAR*D trial. Am J Hum Genet, 78(5):804-814. 2006. [PubMed]
Miller NH, Marosy B, Justice CM, Novak, SM, Tang, EY, Boyce P, Pettengil J, Doheny, KF, Pugh, EW, Wilson AF. Linkage analysis of genetic loci for kyphoscoliosis on chromosomes 5p13, 13q13.3 and 13q32. Am J Med Genet, 140A(10):1059-1068. 2006. [PubMed]
Roy-Gagnon M-H, Mathias RA, Fallin MD, Jee SH, Broman, KW, Wilson AF. An extension of the regression of Offspring on Mid-Parent method to test for association and estimate locus-specific heritability: The revised ROMP method. Ann Hum Genet, 72:115-25. 2008. [PubMed]
Herrera-Galeano JE, Becker DM, Wilson AF, Yanek LR, Bray P, Vaidya D, Faraday N, Becker LC. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arterioscler Thromb Vasc Biol, 28:1484-90. 2008. [PubMed]
Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, Bouffard GG, Chines PS, Cruz P, Hansen NF, Teer JK, Maskeri B, Young AC, Manolio TA, Wilson AF, Finkel T, Hwang P, Arai A, Remaley AT, Sachdev V, Shamburek R, Cannon RO, Green ED, NISC Comparative Sequencing Program. The ClinSeq Project: Piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 19(9):1665-1674. 2009. [PubMed]
Sung H, Kim Y, Cai J, Cropp CD, Simpson CL, Li Q, Perry BC, Sorant AJM, Bailey-Wilson JE, Wilson AF. Comparison of results from tests of association in unrelated individuals with uncollapsed and collapsed sequence variants using tiled regression. BMC Proc, 5(Suppl 9): S15. 2011.
Wilson AF, Blangero J, Ziegler A. Lessons learned from Genetic Analysis Workshop 17: Transitioning from genome-wide associations studies to whole-genome statistical genetic analysis. Genet Epidemiol, 35(Suppl 8):S107-14. 2011.[PubMed]
Bailey-Wilson, Wilson AF. Linkage analysis in the next generation sequencing era. Hum Hered, 72(4):228-36. 2011. [PubMed]
Suktitipat B, Mathias RA, Vaidya D, Yanek LR, Young JH, Becker LC, Becker DM, Wilson AF, Fallin MD. The robustness of generalized estimating equations for association tests in extended family data. Hum Hered, 74(1):17-26. 2012. [PubMed]
Justice CM, Yagnik G, Kim Y, Peter I, Jabs EW, Erazo M, Ye X, Ainehsazan E, Shi L, Cunningham ML, Kimonis V, Roscioli T, Wall SA, Wilkie AO, Stoler J, Richtsmeier JT, Heuze Y, Sanchez-Lara PA, Buckley MF, Druschel CM, Mills JL, Caggana M, Romitti PA, Kay DM, Senders C, Taub PJ, Klein OD, Boggan J, Zwienenberg-Lee M, Naydenov C, Kim J, Wilson AF, Boyadjiev SA. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat Genet, 44(12):1360-1364. 2012. [PubMed]
Kirino Y, Zhou Q, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, Ozyazgan Y, Ugurlu S, Erer B, Abaci N, Ustek D, Meguro A, Ueda A, Takeno M, Inoko H, Ombrello MJ, Satorius CL, Maskeri B, Mullikin JC, Sun HW, Gutierrez-Cruz G, Kim Y, Wilson AF, Kastner DL, Gul A, Remmers EF. Targeted resequencing implicates the familial Mediterranean fever gene MEFV and the toll-like receptor 4 gene TLR4 in Behcet disease. Proc Natl Acad Sci USA, 110(20):8134-8139. 2013. [PubMed]
Pemov A, Sung H, Hyland PL, Sloan JL, Ruppert SL, Baldwin AM, Boland JF, Bass SE, Lee HJ, Jones KM, Zhang X, Mullikin JC, Widemann BC, Wilson AF, Stewart DR. Genetic Modifiers of Neurofibromatosis Type 1-Associated Cafe-au-Lait Macule Count Identified Using Multi-platform Analysis. PLoS Genet, 10(10):e1004575. 2014. [PubMed]
Carter TC, Pangilinan F, Molloy AM, Fan R, Wang Y, Shane B, Gibney ER, Midttun O, Ueland PM, Cropp CD, Kim Y, Wilson AF, Bailey-Wilson JE, Brody LC, Mills JL. Common Variants at Putative Regulatory Sites of the Tissue Nonspecific Alkaline Phosphatase Gene Influence Circulating Pyridoxal 5'-Phosphate Concentration in Healthy Adults. J Nutr Jul, 145(7):1386-1393. 2015. [PubMed]
-
Publications
Wilson AF, Cohen JC. Hypotheses for testing deviations from random integration: evidence for nonrandom retroviral integration. Genomics, 3(2):137-142. 2008. [PubMed]
Amos CI, Elston RC, Wilson AF, Bailey-Wilson JE. A more powerful robust sib-pair test of linkage for quantitative traits. Genet Epidemiol, 6(3):435-449. 1989. [PubMed]
Wilson AF, Elston RC, Sellers TA, Bailey-Wilson JE, Gersting JM, Deen DK, Sorant AJM, Tran LD, Amos CI, Siervogel RM. Stepwise oligogenic segregation and linkage analysis illustrated with dopamine-beta-hydroxylase activity. Am J Med Genet, 35:425-432. 1990. [PubMed]
Wilson AF, Elston RC, Tran LD, Siervogel RM. Use of the robust sib-pair method to screen for single-locus, multiple-locus, and pleiotropic effects: application to traits related to hypertension. Am J Hum Genet, 48:862-872. 1991. [PubMed]
Wilson AF, Elston RC. Statistical validity of the Haseman-Elston sib-pair test in small samples. Genet Epidemiol, 10(6):593-598. 1993. [PubMed]
Goldin LR, Chase GA, Wilson AF. Regional inference with averaged P values increases the power to detect linkage. Genet Epidemiol, 17(3):157-164. 1999. [PubMed]
Wilson AF, Sorant AJ. Equivalence of single- and multilocus markers: power to detect linkage with composite markers derived from biallelic loci. Am J Hum Genet, 66(5):1610-1615. 2000. [PubMed]
Miller NH, Justice CM, Marosy B, Doheny KF, Pugh E, Zhang J, Wilson AF. Identification of candidate regions in familial idiopathic scoliosis. Spine 30(10):1181-1187. 2005. [PubMed]
McMahon FJ, Buervenich S, Charney D, Lipsky R, Rush AJ, Wilson AF, Sorant AJM, Papanicolaou GJ, Laje G, Fava M, Trivedi M, Wisniewski S, Manji H. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of citalopram treatment: Results from the STAR*D trial. Am J Hum Genet, 78(5):804-814. 2006. [PubMed]
Miller NH, Marosy B, Justice CM, Novak, SM, Tang, EY, Boyce P, Pettengil J, Doheny, KF, Pugh, EW, Wilson AF. Linkage analysis of genetic loci for kyphoscoliosis on chromosomes 5p13, 13q13.3 and 13q32. Am J Med Genet, 140A(10):1059-1068. 2006. [PubMed]
Roy-Gagnon M-H, Mathias RA, Fallin MD, Jee SH, Broman, KW, Wilson AF. An extension of the regression of Offspring on Mid-Parent method to test for association and estimate locus-specific heritability: The revised ROMP method. Ann Hum Genet, 72:115-25. 2008. [PubMed]
Herrera-Galeano JE, Becker DM, Wilson AF, Yanek LR, Bray P, Vaidya D, Faraday N, Becker LC. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arterioscler Thromb Vasc Biol, 28:1484-90. 2008. [PubMed]
Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, Bouffard GG, Chines PS, Cruz P, Hansen NF, Teer JK, Maskeri B, Young AC, Manolio TA, Wilson AF, Finkel T, Hwang P, Arai A, Remaley AT, Sachdev V, Shamburek R, Cannon RO, Green ED, NISC Comparative Sequencing Program. The ClinSeq Project: Piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 19(9):1665-1674. 2009. [PubMed]
Sung H, Kim Y, Cai J, Cropp CD, Simpson CL, Li Q, Perry BC, Sorant AJM, Bailey-Wilson JE, Wilson AF. Comparison of results from tests of association in unrelated individuals with uncollapsed and collapsed sequence variants using tiled regression. BMC Proc, 5(Suppl 9): S15. 2011.
Wilson AF, Blangero J, Ziegler A. Lessons learned from Genetic Analysis Workshop 17: Transitioning from genome-wide associations studies to whole-genome statistical genetic analysis. Genet Epidemiol, 35(Suppl 8):S107-14. 2011.[PubMed]
Bailey-Wilson, Wilson AF. Linkage analysis in the next generation sequencing era. Hum Hered, 72(4):228-36. 2011. [PubMed]
Suktitipat B, Mathias RA, Vaidya D, Yanek LR, Young JH, Becker LC, Becker DM, Wilson AF, Fallin MD. The robustness of generalized estimating equations for association tests in extended family data. Hum Hered, 74(1):17-26. 2012. [PubMed]
Justice CM, Yagnik G, Kim Y, Peter I, Jabs EW, Erazo M, Ye X, Ainehsazan E, Shi L, Cunningham ML, Kimonis V, Roscioli T, Wall SA, Wilkie AO, Stoler J, Richtsmeier JT, Heuze Y, Sanchez-Lara PA, Buckley MF, Druschel CM, Mills JL, Caggana M, Romitti PA, Kay DM, Senders C, Taub PJ, Klein OD, Boggan J, Zwienenberg-Lee M, Naydenov C, Kim J, Wilson AF, Boyadjiev SA. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat Genet, 44(12):1360-1364. 2012. [PubMed]
Kirino Y, Zhou Q, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, Ozyazgan Y, Ugurlu S, Erer B, Abaci N, Ustek D, Meguro A, Ueda A, Takeno M, Inoko H, Ombrello MJ, Satorius CL, Maskeri B, Mullikin JC, Sun HW, Gutierrez-Cruz G, Kim Y, Wilson AF, Kastner DL, Gul A, Remmers EF. Targeted resequencing implicates the familial Mediterranean fever gene MEFV and the toll-like receptor 4 gene TLR4 in Behcet disease. Proc Natl Acad Sci USA, 110(20):8134-8139. 2013. [PubMed]
Pemov A, Sung H, Hyland PL, Sloan JL, Ruppert SL, Baldwin AM, Boland JF, Bass SE, Lee HJ, Jones KM, Zhang X, Mullikin JC, Widemann BC, Wilson AF, Stewart DR. Genetic Modifiers of Neurofibromatosis Type 1-Associated Cafe-au-Lait Macule Count Identified Using Multi-platform Analysis. PLoS Genet, 10(10):e1004575. 2014. [PubMed]
Carter TC, Pangilinan F, Molloy AM, Fan R, Wang Y, Shane B, Gibney ER, Midttun O, Ueland PM, Cropp CD, Kim Y, Wilson AF, Bailey-Wilson JE, Brody LC, Mills JL. Common Variants at Putative Regulatory Sites of the Tissue Nonspecific Alkaline Phosphatase Gene Influence Circulating Pyridoxal 5'-Phosphate Concentration in Healthy Adults. J Nutr Jul, 145(7):1386-1393. 2015. [PubMed]
Last updated: June 21, 2022
