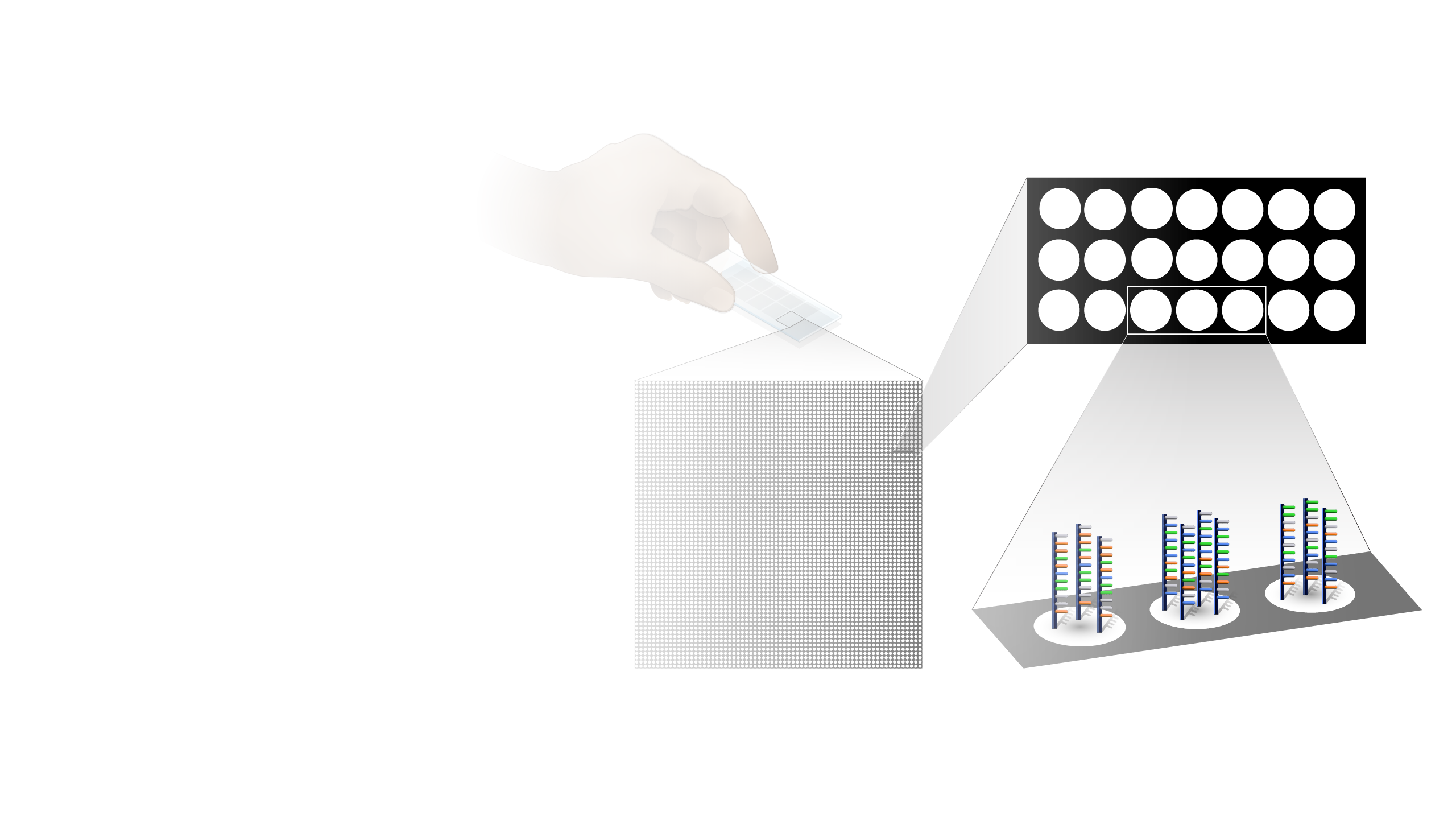
Microarray Technology
Definition
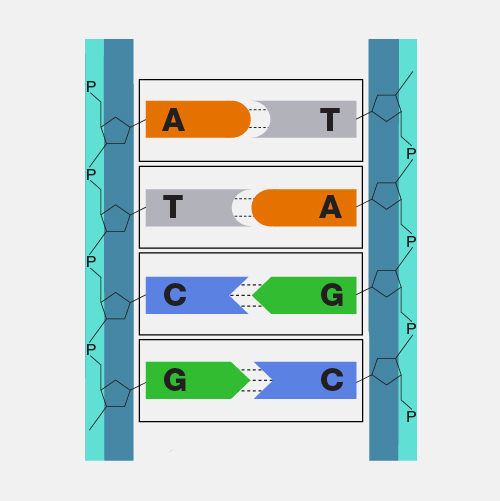
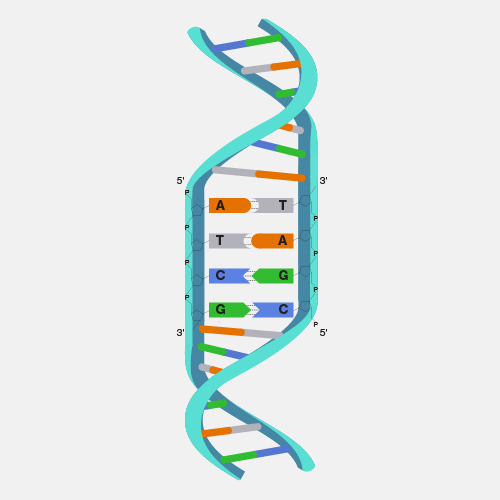
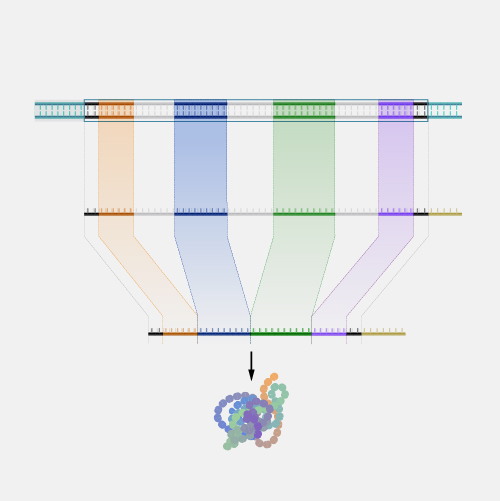
Microarray technology is a general laboratory approach that involves binding an array of thousands to millions of known nucleic acid fragments to a solid surface, referred to as a “chip.” The chip is then bathed with DNA or RNA isolated from a study sample (such as cells or tissue). Complementary base pairing between the sample and the chip-immobilized fragments produces light through fluorescence that can be detected using a specialized machine. Microarray technology can be used for a variety of purposes in research and clinical studies, such as measuring gene expression and detecting specific DNA sequences (e.g., single-nucleotide polymorphisms, or SNPs ).
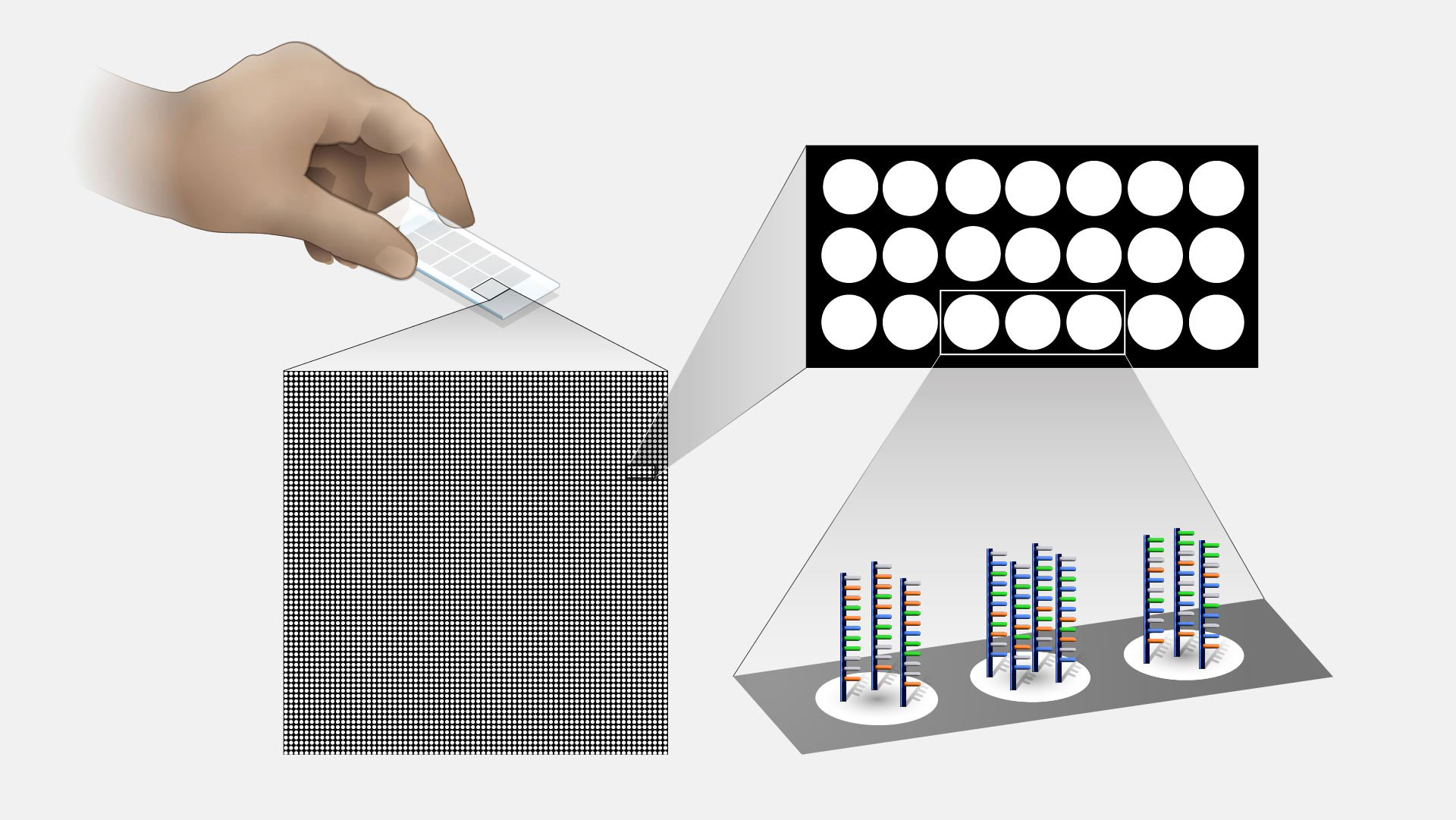
Narration
Microarray technology. Microarrays were revolutionary. They really allow genomic analysis without sequencing, which tremendously reduced the cost of doing large studies across a wide area of biology and biomedicine. Two things that you were able to do there. On the one hand, you're able to look at gene expression or the amount of gene product, RNA, from any given gene that you found in a cell. And the second thing you were able to look at easily was single nucleotide polymorphisms, or SNPs, which were useful for genome-wide association studies, or GWASs. Both of those approaches were used across all of the major human diseases, a large number of less common human diseases, and to also in our model organisms and in other organisms in this world of ours.