Neil A. Hanchard, M.B.B.S., D.Phil.
Childhood Complex Disease Genomics Section
Childhood Complex Disease Genomics Section
M.B.B.S., University Of The West Indies
D.Phil., University Of Oxford
Pediatric Residency, Mayo Clinic, Minnesota
Medical Genetics Residency, Baylor College of Medicine, Houston, Texas
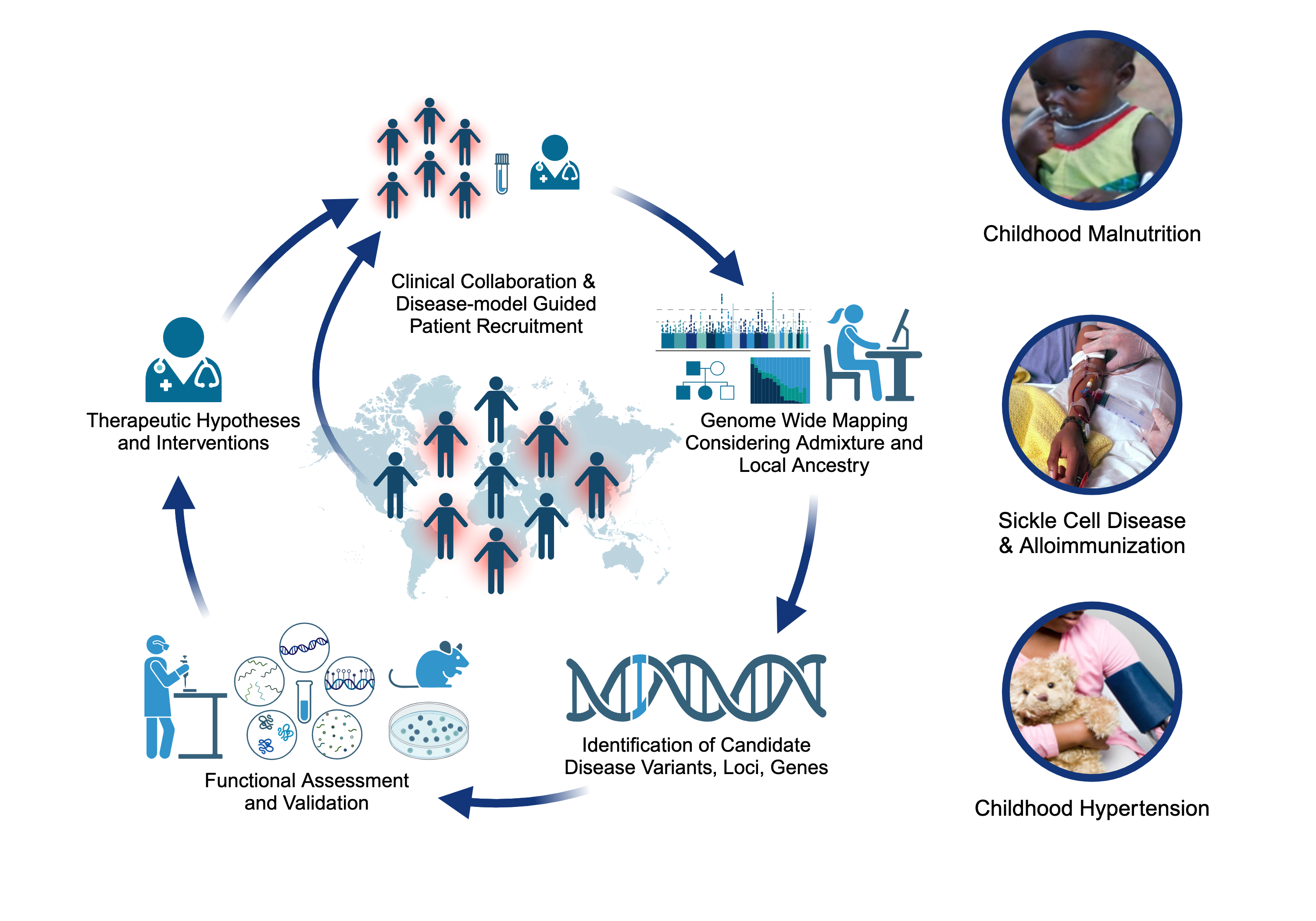
Projects
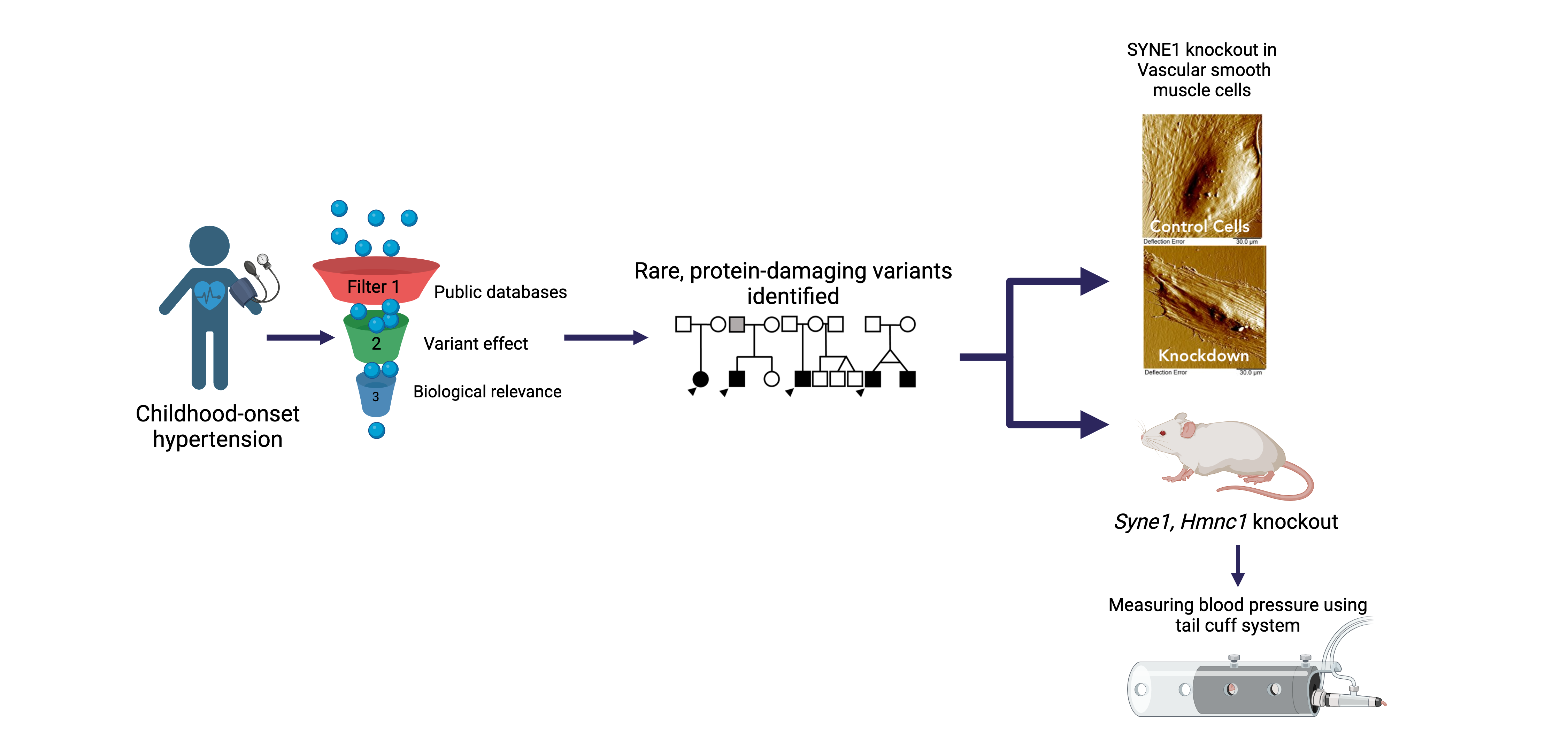
Childhood-onset Essential Hypertension (COEH)
Childhood-onset essential hypertension – in which there is elevated blood pressure in childhood without an obvious cause - affects 1-2% of all children, accounting for about 40% of all hypertension diagnoses in children, and is found more often among populations of African- or Hispanic ancestries. Children with COEH are difficult to diagnose, but tend to have a strong family history of hypertension. Although a role for genetics in the development of the disease has been supported, it is unclear whether this genetic risk mirrors that observed in adults (multiple genes all with small effects) or is more consistent with rare Mendelian diseases that are more striking in childhood (single genes with large effects). Crucially, the treatment of CEOH mirrors that of adults, partly because the underlying pathophysiology is assumed to be the same. We are exploring a model of Mendelian and rare variant contributions to COEH through cohort-based and natural-history gene-discovery coupled with functional characterization and consideration of existing pharmaceutical interventions [PMID: 38716726]. Our work includes working alongside national and international collaborators to expand our COEH cohort and better characterize the vascular and clinical phenotype of gene variant carriers.
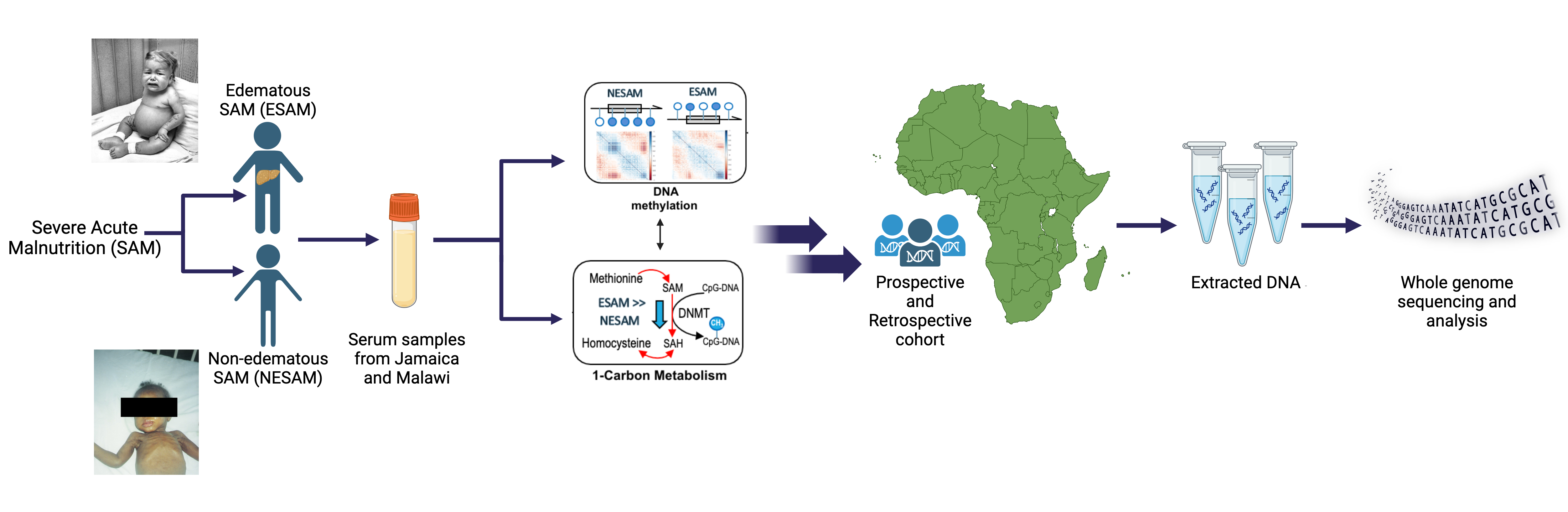
Severe Acute Malnutrition (SAM)
Approximately two million children worldwide under the age of five experience severe malnutrition annually, with an estimated 20-30% not surviving. Classically, severe acute malnutrition (SAM) manifests in one of two forms: the non-edematous form (NESAM or marasmus) which is characterized by chronic wasting, and the edematous form (ESAM, kwashiorkor, or marasmic-kwashiorkor), characterized by more severe multisystem dysfunction. Despite decades of research, the underlying reasons why some children develop ESAM versus NESAM remain elusive, as the etiology of molecular differences in the pathophysiology underlying to two forms being largely unexplored. Our research employs a multi-'omics' approach, focusing on cohorts from diverse ancestries residing in regions where ESAM and SAM are prevalent [PMID: 31857576]. Within our laboratory, we are constructing a functional hepatocyte model to simulate fatty liver in ESAM cases, alongside identifying single nucleotide polymorphisms within ESAM and NESAM samples obtained from Jamaica and Malawi. Additionally, we are fostering international collaborations to establish a comprehensive repository of SAM samples, collaborating with researchers in SAM high-incidence countries to create a Kwashiorkor Collaborative Network (KwashNet). Through this we hope to better understand the etiologic and pathophysiologic differences between ESAM and NESAM. The findings of this study will be used to assist in the development of targeted interventions designed to correct the unique metabolic disturbances of kwashiorkor.
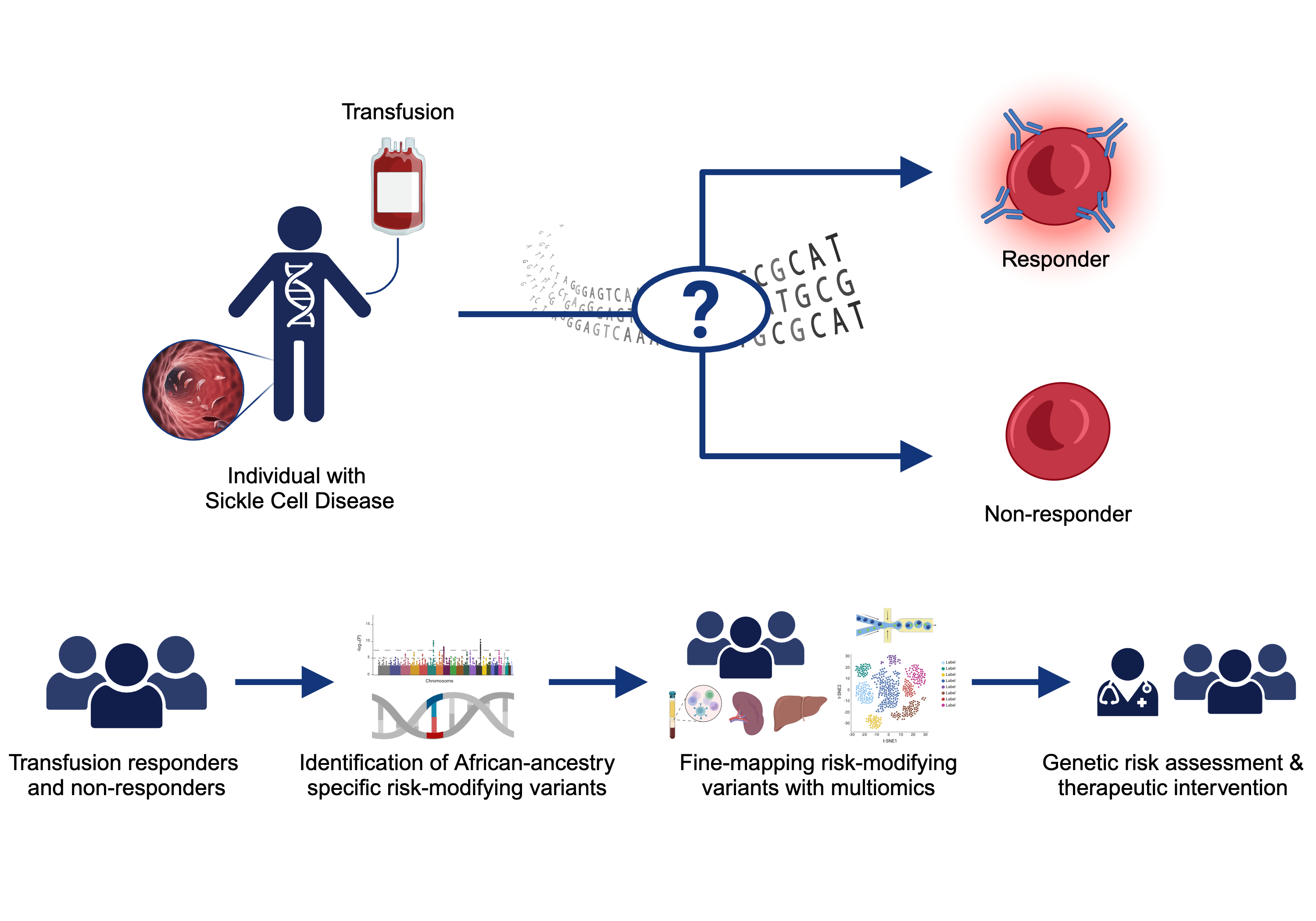
Sickle Cell Disease Alloimmunization
Sickle cell disease (SCD) is one of the most common monogenetic diseases in the world, with high mortality rates and a lowered life expectancy. Regular transfusion of red blood cells (RBCs) is a mainstay of treatment; however, repeated transfusions increase the risk of the recipent developing antibodies against donor RBCs (known as alloimmunization). RBC alloimmunization is seen in up to 20% of multiply-transfused SCD individuals, with some studies reporting rates twice as high. In the most severe cases, transfused patients develop alloantibodies with almost every new transfusion – so called transfusion ‘super-responders’; this creates a major challenge to find compatible blood for future transfusions. Fundamentally, the reason some individuals develop alloantibodies when most do not, remains unclear. In our previous studies, we identified a genome-wide significant association between an intergenic locus on chromosome 5q33 and being a transfusion responder and replicated this in a second cohort [PMID: 30578281]. We are utilizing a multiomics approach to fine map the functional consequence(s) of genetic variation our risk locus. We are also applying trans- and local-ancestry admixture mapping models to other cohorts of alloimmunization in sickle cell disease as a means of identifying novel risk alleles and refining the genetic risk for alloimmunization. Collectively, these efforts seek to provide insight to the immunological mechanisms underlying the increased alloimmunization risk, and in so doing, advance much needed therapeutic and diagnostic targets to improve the safety of this important clinical intervention.
Childhood Complex Disease Genomics Section Staff

- Technical Lab Manager
- Childhood Complex Disease Genomics Section

- Geneticist (Counselor)
- Childhood Complex Disease Genomics Section

- Staff Scientist
- Childhood Complex Disease Genomics Section

- Staff Scientist
- Childhood Complex Disease Genomics Section
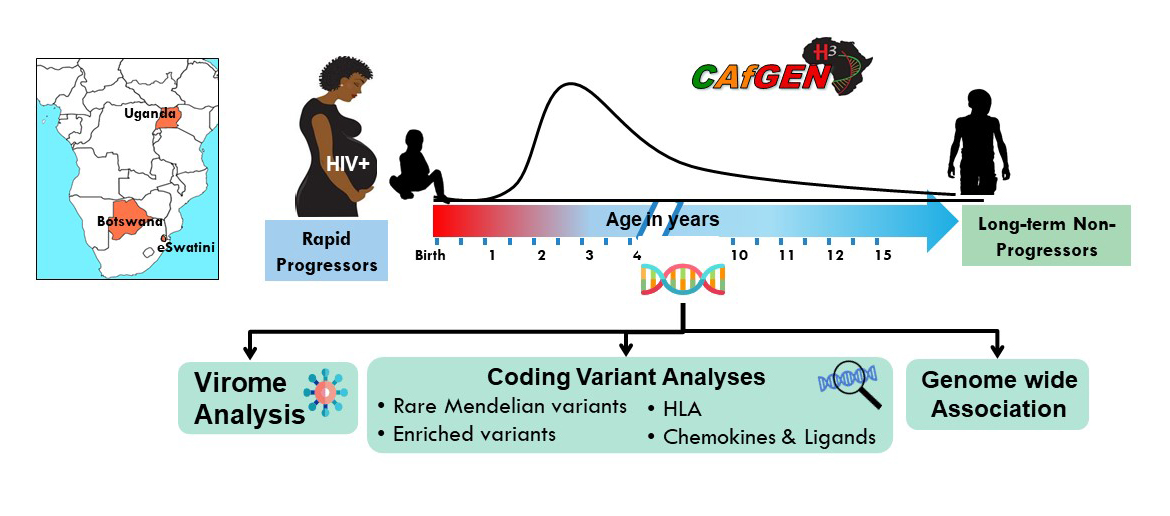
CAfGEN Graduate Students

- CAfGEN Fellow
- University of Botswana

- CAfGEN Fellow
- University of Botswana

- CafGEN Fellow
- University of Botswana

- CAfGEN Fellow
- Makerere College of Health Sciences
Fellows
- Predoctoral IRTA Fellow, CAfGEN Fellow
- Childhood Complex Disease Genomics Section

- Postbaccalaureate Fellow
- Childhood Complex Disease Genomics Section

- Predoctoral IRTA Fellow
- Childhood Complex Disease Genomics Section

- Postbaccalaureate Fellow
- Childhood Complex Disease Genomics Section

- Postbaccalaureate Fellow
- Childhood Complex Disease Genomics Section

- Postbaccalaureate Fellow
- Childhood Complex Disease Genomics Section

- Postbaccalaureate Fellow
- Childhood Complex Disease Genomics Section

- Postdoctoral Fellow
- Childhood Complex Disease Genomics Section

- Postdoctoral Fellow
- Childhood Complex Disease Genomics Section
Last updated: February 12, 2025