Last updated: July 03, 2014
2013 News Feature Nanopore Dna Sequencing New Approaches To An Old Challenge
Nanopore DNA sequencing: New approaches to an old challenge
In the fast-paced world of DNA sequencing, nanopores are all the rage
By Steven Benowitz
Associate Director of Communications, Extramural Research Program
In 2012, MIT Technology Review named nanopore sequencing one of its "10 breakthrough technologies" of the year. Using nanopore technology to sequence DNA, it said, "could make genome sequencing a routine medical procedure." A leading company in the field, UK-based Oxford Nanopore Technologies, announced last year its intention to begin selling a disposable, nanopore-based gene sequencing device the size of a USB memory stick that could plug into a laptop computer. While Oxford has yet to commercialize its product, several other companies have jumped into the nanopore game.
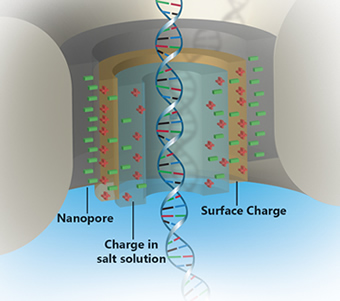
Nanopore-based DNA sequencing involves threading single DNA strands through tiny pores in a membrane. Bases - the chemical letters of DNA - are read one at a time as they squeeze through the nanopore. The bases are identified by measuring differences in their effect on ions and electrical current flowing through the pore. Nanopores used in DNA sequencing are very tiny, perhaps only about 2 nanometers in diameter. A human hair, in contrast, is 100,000 nanometers wide.
Using nanopores to sequence DNA offers many potential advantages over current methods. It costs less, and can be done faster, in real time. The same molecule can be studied over and over again. Most current techniques are more complicated than that. DNA has to be isolated, chemically labeled and copied. It has to be broken up, and small segments have to be sequenced many times.
A fast and inexpensive method of sequencing DNA could have a dramatic effect on clinical care and research. Clinicians, for example, could use this information to help determine a person's risk for a specific disease or condition, or the best choice of a drug.
At the National Human Genome Research Institute (NHGRI), Jeffery A. Schloss, Ph.D., program director for the Genome Technology Program, which administers Advanced Sequencing Technology grants, sees vast potential in using nanopores for DNA sequencing.
"Reading DNA directly is very attractive for a number of reasons, particularly because it can eliminate so much processing time, and may allow us to accurately read long sections of DNA at once," said Dr. Schloss, who is also director of the NHGRI Division of Genome Sciences.
NHGRI has devoted considerable resources to developing nanopore technologies, including five new grants in September 2013. It's not the first time the program, which began in 2004, has awarded grants to researchers in the nanopore technology field.
Recently, researchers from Boston University and Technion University in Israel, led by NHGRI grantee Amit Meller, Ph.D., put a new twist on solving an old problem with nanopores. The rapid speed at which DNA strands pass through the tiny holes makes distinguishing bases more difficult. They showed that shining a certain wavelength of light could slow the flow of DNA through synthetic nanopores, potentially making it easier to read the four bases that make up each molecule.
Reporting in the November 2013 issue of Nature Nanotechnology, Dr. Meller's group found that by shining a low-power green laser on a synthetic nanopore made of a thin layer of silicon nitride, it was possible to increase the electric charge near the walls of the pore, which is immersed in salt water. As the current increases, positive ions drag water molecules in the opposite direction of incoming DNA, acting as a brake and slowing its passage through the pore. As a result, nanoscale sensors in the pore would be more accurately able to read each nucleotide going into the pore.
"By switching the laser on and off at will, or increasing or lowering the intensity, we can modify the water flow and slow down the DNA motion, making it easier to read the DNA coming through the nanopores in real time," explained Dr. Meller, who is an associate professor of biomedical engineering at Boston University.
Most of the current DNA-sequencing approaches with nanopores use an enzyme placed at the entrance to a protein pore to control the rate by which DNA passes through. This is necessary to give the sensor enough time to distinguish between the bases. Other groups are trying to use synthetic pores and an enzyme to slow the DNA.
"Dr. Meller's group's observations might allow the use of light to dynamically control this rate through a synthetic pore. If this technique holds up, it could be a very big step to solving this problem," said Dr. Schloss.
A work in progress
Because using nanopores to sequence DNA might be less complicated and more direct than current methods, the DNA-sequencing device may become smaller as well, eventually reaching the size of a smartphone, Dr. Meller said.
The use of nanopores in DNA sequencing has other advantages as well.
Two research teams at the University of California, Santa Cruz and the University of Washington recently showed that nanopores made of protein can also identify specific epigenetic changes, modifications that occur over the lifetime of the DNA within cells. This has only been possible by introducing numerous biochemical processing steps with routine DNA-sequencing methods. These new studies showed that those steps could be avoided by using nanopores. Understanding such epigenetic changes may help provide new insights into the development of disease and new therapies.
One vexing problem with current DNA-sequencing techniques is the "read length," or the number of consecutive base pairs that can be distinguished. Using nanopores, long stretches of DNA can be zipped back and forth through the pore and can be read several times. Each "read" might be thousands of bases long - instead of hundreds - which could mean fewer computations, fewer genes missed, greater accuracy and less DNA-sequencing time.
Nanopore techniques could also make it easier for scientists to uncover changes in the DNA - translocations, for example, which involve sections of DNA jumping from one chromosome to another, and copy number variations, where DNA sequences are repeated over and over. These types of changes often play roles in cancer and other diseases, and are hard to detect with current methods.
The nanopore field has two parallel tracks of development: protein and synthetic. Of the two types, protein pores are further along in development. Protein nanopores appear to be better at recognizing nucleotides, while synthetic pores should last longer and can be manufactured on a large scale. They can also be integrated into electronic devices using the same, well developed methods that are used to make computer chips.
While Dr. Meller's team is using light to slow the progress of the DNA through synthetic nanopores, a number of investigators are using a polymerase, a molecular motor that drags DNA through the pores, one base at a time. Several companies are close to commercializing the first nanopore DNA sequencing approaches involving protein nanopores and a polymerase.
"NHGRI has invested in nanopore technology because it shows great promise," said Dr. Schloss. "It's a relatively young field, and there's still a long way to go to show that nanopores will ultimately be effective for DNA sequencing."
Last Updated: July 3, 2014