Last updated: November 21, 2013
New NIH awards focus on nanopore technology for DNA sequencing

National Human Genome Research Institute (NHGRI)
www.genome.gov
New NIH awards focus on nanopore technology for DNA sequencing
Bethesda, Md., Fri., Sept. 6, 2013 - The use of nanopore technology aimed at more accurate and efficient DNA sequencing is the main focus of grants awarded by the National Institutes of Health. The grants - nearly $17 million to eight research teams - are the latest awarded through the National Human Genome Research Institute (NHGRI)'s Advanced DNA Sequencing Technology program, which was launched in 2004. NHGRI is part of NIH.
"Nanopore technology shows great promise, but it is still a new area of science. We have much to learn about how nanopores can work effectively as a DNA sequencing technology, which is why five of the program's eight grants are exploring this approach," said Jeffery A. Schloss, Ph.D., program director for NHGRI's Advanced DNA Sequencing Technology program and director of the Division of Genome Sciences.
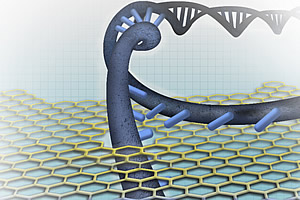
Nanopore-based DNA sequencing involves threading single DNA strands through tiny pores. Individual base pairs - the chemical letters of DNA - are then read one at a time as they pass through the nanopore. The bases are identified by measuring the difference in their effect on current flowing through the pore. For perspective, a human hair is 100,000 nanometers in diameter; a strand of DNA is only 2 nanometers in diameter. Nanopores used in DNA sequencing are 1 to 2 nanometers in diameter.
This technology offers many potential advantages over current DNA sequencing methods, said Dr. Schloss. Such advantages include real-time sequencing of single DNA molecules at low cost and the ability for the same molecule to be reassessed over and over again. Current systems involve isolating DNA and chemically labeling and copying it. DNA has to be broken up, and small segments are sequenced many times. Only the first step of isolating the DNA would be necessary with nanopore technology.
Innovation is crucial in these as well as the other (non-nanopore) studies being funded. For example, one research team eventually hopes to use light to sequence DNA on a cell phone camera chip for under $100.
The new grants are awarded to:
- University of Illinois, Urbana-Champaign, $2.47 million over four years (pending available funds)
Principal Investigator: Oleksii Aksimentiev, Ph.D.
Dr. Aksimentiev and his colleagues plan to use nanopores as sensors. The researchers are studying the effects of combining synthetic nanopores with a light-based technique to control the flow of DNA molecules through the pores. They will use a type of spectroscopy to read the chemical sequence of the DNA.
- University of New Mexico Health Sciences Center, Albuquerque, $1.35 million over three years (pending available funds)
Principal Investigator: Jeremy Edwards, Ph.D.
Dr. Edwards and his colleagues plan to develop innovative molecular biology tools to improve whole-genome sequencing, which entails reading a person's entire genetic blueprint. The researchers hope that better methods of preparing the DNA molecules for sequencing will help scientists identify and link genetic variants to disease and, ultimately, lead to new treatments.
- University of Washington, Seattle, $3.83 million over four years (pending available funds)
Principal Investigator: Jens Gundlach, Ph.D.
The researchers plan to continue developing the use of nanopore DNA sequencing technology involving a type of protein nanopore called MspA. Part of their research will focus on improving the control of movement of DNA through the nanopore and on developing algorithms to identify DNA bases.
- Columbia University, New York City, $5.25 million over three years (pending available funds)
Principal Investigators: Jingyue Ju, Ph.D., George M. Church, Ph.D., (Harvard Medical School, Boston) and James John Russo, Ph.D. (Columbia University, New York City)
Dr. Ju and his colleagues plan to develop a miniaturized electronic system using nanopores to analyze single molecules of DNA in real time. They will construct large arrays of nanopores to create DNA sequencing chips, enabling them to determine DNA bases during a specific biochemical reaction. They hope this technique will enable them to read large sections of DNA more accurately and rapidly than is now possible.
- Eve Biomedical, Inc., Mountain View, Calif., $493,000 over two years (pending available funds)
Principal Investigator: Theofilos Kotseroglou, Ph.D.
Dr. Kotseroglou's research team intends to develop a DNA sequencing system that can sequence an entire human genome for under $100. The overall system will be based on using light to sequence DNA on a cell phone camera chip. For now, his group plans to continue studying ways to accurately read long sections of DNA and develop software tools and bioinformatics.
- University of Massachusetts, Amherst, $1.07 million over four years (pending available funds)
Principal Investigator: Murugappan Muthukumar, Ph.D.
Dr. Muthukumar's research group plans a theoretical approach to study several major challenges underlying nanopore-based DNA sequencing, including slowing down the rate at which DNA molecules flow through the pores, the effects of specific ions, changes in the shape of the DNA molecule and other aspects of the environment.
- University of North Carolina at Chapel Hill, $2.05 million over four years (pending available funds)
Principal Investigator: John Michael Ramsey, Ph.D.
Dr. Ramsey and his co-workers plan to develop a low-cost method for rapidly mapping individual genomes. Such maps will help determine how large mutations in DNA structure contribute to human disease and improve diagnostic testing using genomics.
- Electronic Biosciences, Inc., San Diego, $239,000
Principal Investigator: Anna Schibel, Ph.D.
Dr. Schibel and her co-workers will develop chemical methods to slow the rate by which single-stranded DNA molecules pass through protein nanopores. Such approaches may enable the development of faster, lower-cost DNA sequencing techniques.
To read the grant abstracts go to Advanced Sequencing Technology Awards 2013. For more details about the full technology development program, go to: Genome Technology Program.
The costs of DNA sequencing have greatly declined since 2003, when the genome sequencing performed under the Human Genome Project was completed at a cost of approximately $1 billion. Only a year later, in 2004, sequencing a human genome cost an estimated $10-50 million, thanks to improvements in technologies and tools. By 2009, NHGRI met its goal of producing high-quality human genome sequences at a 100-fold reduction in price, or $100,000. While achieving another 100-fold drop in price has been difficult, sequencing a person's genome today costs about $5,000 to $6,000 (http://www.genome.gov/sequencingcosts).
The grant numbers of the awards are the following: R01 HG007406; R01 HG006876; R01 HG005115; R01 HG007415; R43 HG007386; R01 HG002776; R01 HG007407; and R43 HG006878.
NHGRI is one of the 27 institutes and centers at the National Institutes of Health. The NHGRI Extramural Research Program supports grants for research and training and career development at sites nationwide. Additional information about NHGRI can be found at www.genome.gov.
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 institutes and centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit www.nih.gov.
Contact:
NHGRI Communications
Steven Benowitz
(301) 451-8325
Steven.Benowitz@nih.gov