Biography
Dr. David Bodine, Ph.D. is Chief of the Genetics and Molecular Biology Branch and Head of the Hematopoiesis Section at the National Human Genome Research Institute (NHGRI). NHGRI is one of the 27 Institutes and Centers making up the National Institutes of Health (NIH).
Dr. Bodine received his undergraduate degree from Colby College in 1976, a master's degree in Human Genetics from Rutgers in 1977 and a Ph.D. for his work at the Jackson Laboratory through the University of Maine in 1984. After postdoctoral work at NIH's National Heart, Lung and Blood Institute, Dr. Bodine founded the Hematopoiesis Section in 1993 as part of the newly formed Intramural Research Program of NHGRI. In 1995, Dr. Bodine was promoted to Senior Investigator with tenure at NHGRI, and in 2006 was named chief of NHGRI's Genetics and Molecular Biology Branch.
At NIH, Dr. Bodine serves as the Vice Chair and Ombudsman for the trans-NIH Animal Research Advisory Committee. In addition, he is a member of the trans-NIH Research Facility Advisory Committee and the Central Animal Facilities Working Group. Finally, Dr. Bodine serves on the NIH Subcommittee on Sex as a Biological Variable.
Dr. Bodine has won numerous awards during his career. As an undergraduate, he received the Webster Chester Biology prize, and was awarded an honorary degree from Colby College in 2013 for his contributions to the genetics of blood disorders. As a graduate student, he was the recipient of a Jackson Laboratory Fellowship. Dr. Bodine received postdoctoral fellowships from both the NIH and the Cooley's Anemia Foundation. More recently, Dr. Bodine received the Daniella Maria Arturi Foundation Pioneer Award for his research into the causes of DBA. Dr. Bodine was elected to serve as a Councilor by the International Society of Experimental Hematology. The American Society of Gene and Cell Therapy has elected Dr. Bodine to serve as President, Secretary, and as a member of the Advisory Council.
Dr. Bodine was a member of the NIH Hematology I Study Section and continues to serve on the NIH Center for Scientific Review College of Reviewers. Dr. Bodine was a member of the National Science Foundation Eukaryotic Genetics Panel and has served on the Leukemia and Lymphoma Society (LLS) Career Development Award Review Panel. He also served the LLS Medical and Scientific Affairs and Professional Education Committees. Dr. Bodine has served on the Scientific Advisory Boards of the Center for Stem Cell and Regenerative Medicine, the National Disease Research Interchange and the Wisconsin Blood Center.
Dr. Bodine is an active member of the American Society of Hematology (ASH). He was elected to serve on the ASH Executive Committee as a Councillor. Currently he serves on the ASH Committee on Scientific Affairs and the ASH Working Group on the Dynamic Nature of Biomedical Research. Dr. Bodine has chaired the ASH Bridge Grant Program Study Section, the ASH SCD Clinical Trials Network Site Selection Study Section and the ASH Scholar Awards Study Section.
Dr. Bodine recently completed a term as Associate Editor of the journal Blood and has served on the Editorial Boards of Blood, Experimental Hematology, British Journal of Hematology, American Journal of Hematology, Gene Therapy and Molecular Therapy.
Having benefited from outstanding mentoring throughout his career, Dr. Bodine is determined to pay that forward by making the mentoring of trainees, particularly those in his own lab, his highest priority. He has served on the ASH Translational Research Training in Hematology (TRTH) and Clinical Research Training Institute (CRTI) Oversight Subcommittees and is active in the ASH Minority Medical Student Award Program. In recognition for the mentoring of his own trainees, Dr. Bodine has been named NHGRI Mentor of the Year three times, most recently in 2012, and was named NIH Mentor of the Year in 2004.
-
Biography
Dr. David Bodine, Ph.D. is Chief of the Genetics and Molecular Biology Branch and Head of the Hematopoiesis Section at the National Human Genome Research Institute (NHGRI). NHGRI is one of the 27 Institutes and Centers making up the National Institutes of Health (NIH).
Dr. Bodine received his undergraduate degree from Colby College in 1976, a master's degree in Human Genetics from Rutgers in 1977 and a Ph.D. for his work at the Jackson Laboratory through the University of Maine in 1984. After postdoctoral work at NIH's National Heart, Lung and Blood Institute, Dr. Bodine founded the Hematopoiesis Section in 1993 as part of the newly formed Intramural Research Program of NHGRI. In 1995, Dr. Bodine was promoted to Senior Investigator with tenure at NHGRI, and in 2006 was named chief of NHGRI's Genetics and Molecular Biology Branch.
At NIH, Dr. Bodine serves as the Vice Chair and Ombudsman for the trans-NIH Animal Research Advisory Committee. In addition, he is a member of the trans-NIH Research Facility Advisory Committee and the Central Animal Facilities Working Group. Finally, Dr. Bodine serves on the NIH Subcommittee on Sex as a Biological Variable.
Dr. Bodine has won numerous awards during his career. As an undergraduate, he received the Webster Chester Biology prize, and was awarded an honorary degree from Colby College in 2013 for his contributions to the genetics of blood disorders. As a graduate student, he was the recipient of a Jackson Laboratory Fellowship. Dr. Bodine received postdoctoral fellowships from both the NIH and the Cooley's Anemia Foundation. More recently, Dr. Bodine received the Daniella Maria Arturi Foundation Pioneer Award for his research into the causes of DBA. Dr. Bodine was elected to serve as a Councilor by the International Society of Experimental Hematology. The American Society of Gene and Cell Therapy has elected Dr. Bodine to serve as President, Secretary, and as a member of the Advisory Council.
Dr. Bodine was a member of the NIH Hematology I Study Section and continues to serve on the NIH Center for Scientific Review College of Reviewers. Dr. Bodine was a member of the National Science Foundation Eukaryotic Genetics Panel and has served on the Leukemia and Lymphoma Society (LLS) Career Development Award Review Panel. He also served the LLS Medical and Scientific Affairs and Professional Education Committees. Dr. Bodine has served on the Scientific Advisory Boards of the Center for Stem Cell and Regenerative Medicine, the National Disease Research Interchange and the Wisconsin Blood Center.
Dr. Bodine is an active member of the American Society of Hematology (ASH). He was elected to serve on the ASH Executive Committee as a Councillor. Currently he serves on the ASH Committee on Scientific Affairs and the ASH Working Group on the Dynamic Nature of Biomedical Research. Dr. Bodine has chaired the ASH Bridge Grant Program Study Section, the ASH SCD Clinical Trials Network Site Selection Study Section and the ASH Scholar Awards Study Section.
Dr. Bodine recently completed a term as Associate Editor of the journal Blood and has served on the Editorial Boards of Blood, Experimental Hematology, British Journal of Hematology, American Journal of Hematology, Gene Therapy and Molecular Therapy.
Having benefited from outstanding mentoring throughout his career, Dr. Bodine is determined to pay that forward by making the mentoring of trainees, particularly those in his own lab, his highest priority. He has served on the ASH Translational Research Training in Hematology (TRTH) and Clinical Research Training Institute (CRTI) Oversight Subcommittees and is active in the ASH Minority Medical Student Award Program. In recognition for the mentoring of his own trainees, Dr. Bodine has been named NHGRI Mentor of the Year three times, most recently in 2012, and was named NIH Mentor of the Year in 2004.
Scientific Summary
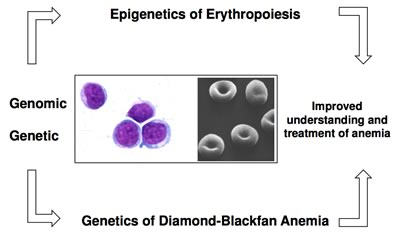
Hematopoiesis is the process of generating all of the different types of blood cells (erythrocytes, lymphocytes, platelets, etc.) circulating throughout the body. All of these cells are derived from a small population of pluripotent hematopoietic stem cells. As pluripotent hematopoietic stem and progenitor cells multiply and differentiate, their progeny lose the ability to differentiate into some lineages and eventually become committed to one single type of cell that ultimately enters the blood circulation. The Hematopoiesis Section's research focuses on erythropoiesis, the regenerative process in which undifferentiated hematopoietic cells differentiate into red blood cells. Perturbations of this process cause a variety of disorders ranging from anemia to hematologic malignancies.
One of the Hematopoiesis Section's research objectives is to understand the epigenetic changes that accompany red cell differentiation, including how the erythroid epigenetic profiles differ from the profiles in cells from other lineages. NHGRI’s ENCyclopedia Of DNA Elements (ENCODE) Project was developed to identify all functional elements in the human genome, including epigenetic marks and sites occupied by transcription factors, primarily focusing on cell lines or cultured cells. The Hematopoiesis Section participates in a consortium known as VISION (Validated Systematic IntegratiON of hematopoietic epigenomes) whose goal is to define the epigenetic changes that occur during erythropoiesis. VISION is designed to extend ENCODE into the study of differentiation of primary hematopoietic stem and progenitor cells.
The hematopoietic system provides a unique opportunity for studying differentiation because flow cytometry can be used to separate primary hematopoietic cells into well-defined populations representing distinct lineages and stages of differentiation. This allows the determination of the epigenetic state (chromatin accessibility, histone modifications, transcription factor binding, DNA methylation and 3D chromatin interactions) of the genome in freshly isolated cell types, which is not possible in other organs. These data can be correlated with transcriptional profiles, to determine the regulatory signature that accompanies differentiation into a specific lineage (i.e. red cells). Our most recent studies have used single cell approaches to identify the earliest progenitor cells for the erythroid and several other several lineages. The long-term goal is to identify the critical regulatory pathways that promote erythroid proliferation and differentiation and to manipulate these with small molecules or compounds that could be used to treat anemia.
The Hematopoiesis Section also studies erythropoiesis through the analysis of a congenital anemia known as Diamond-Blackfan Anemia syndrome (DBA). DBA is an inherited, autosomal dominant disorder associated with failed erythropoiesis and often congenital malformations and a predisposition to cancer. Approximately 70 percent of DBA patients have mutations in genes encoding one of ~25 ribosomal protein (RP) subunits, which result in haploinsufficiency and impaired ribosome function. Because DBA mutations do not always result in severe disease, the lack of a molecular diagnosis in 30% of DBA patients prevents the use of sibling donors for hematopoietic stem cell transplantation, the only curative therapy for DBA. The lack of a molecular diagnosis also complicates family planning for parents and siblings of affected individuals. The Hematopoiesis Section is currently employing genomic technologies to determine whether deletions involving RP genes or their regulatory sequences are the cause of DBA in the 30% of patients without a molecular diagnosis. We have used SNP arrays to show that 15% of DBA patients have large deletions that remove an RP gene. Currently we have used whole genome sequencing to identify smaller deletions and insertions in RP genes in an additional ~10% of DBA patients. Finally, the Hematopoiesis Section is developing reporter cell lines that can be used to screen for drugs that increase the levels of ribosomal proteins, which may offer a new treatment for DBA.
-
Scientific Summary
Hematopoiesis is the process of generating all of the different types of blood cells (erythrocytes, lymphocytes, platelets, etc.) circulating throughout the body. All of these cells are derived from a small population of pluripotent hematopoietic stem cells. As pluripotent hematopoietic stem and progenitor cells multiply and differentiate, their progeny lose the ability to differentiate into some lineages and eventually become committed to one single type of cell that ultimately enters the blood circulation. The Hematopoiesis Section's research focuses on erythropoiesis, the regenerative process in which undifferentiated hematopoietic cells differentiate into red blood cells. Perturbations of this process cause a variety of disorders ranging from anemia to hematologic malignancies.
One of the Hematopoiesis Section's research objectives is to understand the epigenetic changes that accompany red cell differentiation, including how the erythroid epigenetic profiles differ from the profiles in cells from other lineages. NHGRI’s ENCyclopedia Of DNA Elements (ENCODE) Project was developed to identify all functional elements in the human genome, including epigenetic marks and sites occupied by transcription factors, primarily focusing on cell lines or cultured cells. The Hematopoiesis Section participates in a consortium known as VISION (Validated Systematic IntegratiON of hematopoietic epigenomes) whose goal is to define the epigenetic changes that occur during erythropoiesis. VISION is designed to extend ENCODE into the study of differentiation of primary hematopoietic stem and progenitor cells.
The hematopoietic system provides a unique opportunity for studying differentiation because flow cytometry can be used to separate primary hematopoietic cells into well-defined populations representing distinct lineages and stages of differentiation. This allows the determination of the epigenetic state (chromatin accessibility, histone modifications, transcription factor binding, DNA methylation and 3D chromatin interactions) of the genome in freshly isolated cell types, which is not possible in other organs. These data can be correlated with transcriptional profiles, to determine the regulatory signature that accompanies differentiation into a specific lineage (i.e. red cells). Our most recent studies have used single cell approaches to identify the earliest progenitor cells for the erythroid and several other several lineages. The long-term goal is to identify the critical regulatory pathways that promote erythroid proliferation and differentiation and to manipulate these with small molecules or compounds that could be used to treat anemia.
The Hematopoiesis Section also studies erythropoiesis through the analysis of a congenital anemia known as Diamond-Blackfan Anemia syndrome (DBA). DBA is an inherited, autosomal dominant disorder associated with failed erythropoiesis and often congenital malformations and a predisposition to cancer. Approximately 70 percent of DBA patients have mutations in genes encoding one of ~25 ribosomal protein (RP) subunits, which result in haploinsufficiency and impaired ribosome function. Because DBA mutations do not always result in severe disease, the lack of a molecular diagnosis in 30% of DBA patients prevents the use of sibling donors for hematopoietic stem cell transplantation, the only curative therapy for DBA. The lack of a molecular diagnosis also complicates family planning for parents and siblings of affected individuals. The Hematopoiesis Section is currently employing genomic technologies to determine whether deletions involving RP genes or their regulatory sequences are the cause of DBA in the 30% of patients without a molecular diagnosis. We have used SNP arrays to show that 15% of DBA patients have large deletions that remove an RP gene. Currently we have used whole genome sequencing to identify smaller deletions and insertions in RP genes in an additional ~10% of DBA patients. Finally, the Hematopoiesis Section is developing reporter cell lines that can be used to screen for drugs that increase the levels of ribosomal proteins, which may offer a new treatment for DBA.

Publications
Pek RH, Yuan X, Rietzschel N, Zhang J, Jackson LK, Nishibori E, Ribeiro A, Simmons WR, Jagadeesh J, SugimotoH, Alam MZ, Garrett LJ, Haldar M, Ralle M, Phillips J, Bodine DM, Hamza I. Hemozoin produced by mammals confers heme tolerance. eLife. 2019. [In Press]
Zhou S, Giannetto M, DeCourcey J, Kang H, Kang N, Li Y, Zheng S, Zhao H, Simmons WR, Wei HS, Bodine DM, Low PS, Nedergaard M, Wan J. Oxygen tension-mediated erythrocyte membrane interactions regulate cerebral capillary hyperemia. Sci Adv, 5(5):eaaw4466. 2019. [PubMed]
Heuston EF, Keller CA, Lichtenberg J, Giardine B, Anderson SM; NIH Intramural Sequencing Center, Hardison RC, Bodine DM. Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points. Epigenetics Chromatin, 11(1):22. 2018. [PubMed]
O'Brien KA, Farrar JE, Vlachos A, Anderson SM, Tsujiura CA, Lichtenberg J, Blanc L, Atsidaftos E, Elkahloun A, An X, Ellis SR, Lipton JM, Bodine DM. Molecular convergence in ex vivo models of Diamond-Blackfan anemia. Blood, 129(23):3111-3120. 2017. [PubMed]
Psaila B, Barkas N, Iskander D, Roy A, Anderson S, Ashley N, Caputo VS, Lichtenberg J, Loaiza S, Bodine DM, Karadimitris A, Mead AJ, Roberts I. Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol, 17:83. 2016. [PubMed]
Farrar JE, Quarello P, Fisher R, O'Brien KA, Aspesi A, Parrella S, Henson AL, Seidel NE, Atsidaftos E, Prakash S, Bari S, Garelli E, Arceci RJ, Dianzani I, Ramenghi U, Vlachos A, Lipton JM, Bodine DM, Ellis SR. Exploiting pre-rRNA processing in Diamond Blackfan anemia gene discovery and diagnosis. Am J Hematol, 89: 985-91. 2014. [PubMed]
Vlachos A, Farrar JE, Atsidaftos E, Muir E, Narla A, Markello TC, Singh SA, Landowski M, Gazda HT, Blanc L, Liu JM, Ellis SR, Arceci RJ, Ebert BL, Bodine DM, Lipton JM. Diminutive somatic deletions in the 5q region lead to a phenotype atypical of classical 5q- syndrome. Blood, 122: 2487-90. 2013. [PubMed]
Hogart A, Lichtenberg J, Ajay SS, Anderson S; NIH Intramural Sequencing Center, Margulies EH, Bodine DM. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res, 22(8):1407-18. 2012. [PubMed]
Farrar JE, Vlachos A, Atsidaftos E, Carlson-Donohoe H, Markello TC, Arceci RJ, Ellis SR, Lipton JM, Bodine DM. Ribosomal Protein Gene Deletions in DBA. Blood, 118 (26):6943-51. 2011. [PubMed]
Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, Anderson SM; NISC Comparative Sequencing Center, Mullikin JC, Gallagher PG, Hardison RC, Margulies EH, Bodine DM. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 118(17):e139-48. 2011. [PubMed]
Gallagher PG, Steiner LA, Liem RI, Owen AN, Cline AP, Seidel NE, Garrett LJ, Bodine DM. Mutation of a barrier insulator in the human ankyrin-1 gene is associated with hereditary spherocytosis. J Clin Invest, 120:4453-65. 2010. [PubMed]
Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood, 116:2826-35. 2010. [PubMed]
Laflamme K, Owen AN, Devlin EE, Yang MQ, Wong C, Steiner LA, Garrett LJ, Elnitski L, Gallagher PG, Bodine DM. Functional analysis of a novel cis-acting regulatory region within the human ankyrin gene (ANK-1) promoter. Mol Cell Biol, 30 (14):3493-502. 2010. [PubMed]
Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, Gallagher PG, Bodine DM. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Bio, 28 (24):7394-401. 2008. [PubMed]
Nemeth MJ, Topol L, Kirby M, Yang Y, Bodine DM. Wnt5a Inhibits Canonical Wnt Signaling in Hematopoietic Stem Cells and Enhances Repopulation. PNAS, 104:15436-41. 2007. [PubMed]
Nemeth M, Kirby M, Bodine DM. Hmgb3 Regulates the Balance Between Hematopoietic Stem Cell Self-Renewal and Differentiation. PNAS, 103:13783-8. 2006. [PubMed]
Broxmeyer HE, Srour EF, Hangoc G, Cooper S, Anderson SM, Bodine DM. High Efficiency Recovery of Hematopoietic Progenitor Cells with Extensive Proliferative and Ex Vivo Expansion Activity and of Hematopoietic Stem Cells with NOD/SCID Mouse Repopulating Ability from Human Cord Blood Cryopreserved for 15 years. PNAS, 100:645-50. 2003. [PubMed]
Sabatino DE, Seidel NS, Aviles-Mendoza GJ, Cline AP, Anderson SM, Gallagher PG, Bodine DM. Long term expression of g-globin mRNA in mouse erythrocytes from retrovirus vectors containing the human g-globin gene fused to the Ank-1 promoter. PNAS, 97:13294-9. 2000. [PubMed]
Dunbar CE, Seidel NE, Doren S, Sellers S, Cline AP, Metzger ME, Agricola BA, Donahue RE, Bodine DM. Improved retroviral gene transfer into murine and Rhesus peripheral blood or bone marrow repopulating cells primed in vivo with Stem Cell Factor and Granulocyte Colony Stimulating Factor. PNAS, 93:11871-76. 1996. [PubMed]
Orlic D, Girard LJ, Jordan CT, Anderson SM, Cline AP, Bodine DM. The level of mRNA encoding amphotropic retrovirus receptor in mouse and human hematopoietic stem cells is low and correlates with the efficiency of retrovirus transduction. PNAS, 93:11097-11012. 1996. [PubMed]
Orlic D, Anderson SM, Biesecker LG, Sorrentino BP, Bodine DM. Pluripotent hematopoietic stem cells contain high levels of mRNA for c-kit, GATA-2, p45NF-E2 and c-myb, and low levels or no mRNA for c-fms and the receptors for granulocyte-colony stimulating factor and interleukins 5 and 7. PNAS, 92:4601-4605. 1995. [PubMed]
Bodine DM, McDonagh KT, Brandt SJ, Ney PA, Agricola B, Byrne E, Nienhuis AW. Development of a high-titer retrovirus producer cell line capable of gene transfer into Rhesus hematopoietic stem cells. PNAS, 87:3738-3742. 1990. [PubMed]
Bodine DM, Karlsson S, and Nienhuis AW. The combination of Interleukin 3 and 6 preserves stem cell function in culture and enhances retrovirus-mediated gene transfer into hematopoietic stem cells. PNAS, 86:8897-8901. 1989. [PubMed]
Bodine DM and Ley TJ. An enhancer element lies 3' to the human A gamma globin gene. EMBO J, 6:2997-3004. 1987. [PubMed]
Bodine DM, Birkenmeier CS, and Barker JE. Spectrin deficient inherited hemolytic anemias in the mouse: Characterization by spectrin synthesis and mRNA activity in reticulocytes. Cell, 37:721-729. 1984. [PubMed]
Last updated: November 18, 2022
