
Adebowale A. Adeyemo, M.D.
Center For Research On Genomics and Global Health
Center for Genomics and Data Science Research
M.D. [MBBS] University of Ibadan, 1987
Biography
Dr. Adeyemo qualified in medicine at the University of Ibadan in Nigeria. After a residency in pediatrics and genetics, he became a faculty member of the College of Medicine, University of Ibadan, Nigeria and a consultant pediatrician/geneticist at the University College Hospital, Ibadan, Nigeria. He subsequently held fellowships in genetic epidemiology and in medical education. He moved to Howard University, Washington, D.C. in 2003 to work in genetic epidemiology at the National Human Genome Center at Howard University. He came to the NIH in 2008 as a Staff Scientist. He became an associate investigator at NHGRI in 2016.
Dr. Adeyemo works on the genomics of complex disease, focusing on cardiometabolic disorders and complex disorders of childhood. He has published widely in genetics and genetic epidemiology. He was the first author of the papers describing: the first genome scan for obesity in an African population, the first genome-wide linkage analysis for serum lipids in an African population, the first study of genetic structure in West Africans using genome-wide markers and the first genome-wide association study (GWAS) for hypertension and blood pressure in African Americans.
Dr. Adeyemo is currently co-chair of the H3Africa Genome Analysis Working Group and serves on the H3ABioNet Scientific Advisory Board. He is a co-creator of the NHGRI electronic atlas of birth defects for diverse populations.
-
Biography
Dr. Adeyemo qualified in medicine at the University of Ibadan in Nigeria. After a residency in pediatrics and genetics, he became a faculty member of the College of Medicine, University of Ibadan, Nigeria and a consultant pediatrician/geneticist at the University College Hospital, Ibadan, Nigeria. He subsequently held fellowships in genetic epidemiology and in medical education. He moved to Howard University, Washington, D.C. in 2003 to work in genetic epidemiology at the National Human Genome Center at Howard University. He came to the NIH in 2008 as a Staff Scientist. He became an associate investigator at NHGRI in 2016.
Dr. Adeyemo works on the genomics of complex disease, focusing on cardiometabolic disorders and complex disorders of childhood. He has published widely in genetics and genetic epidemiology. He was the first author of the papers describing: the first genome scan for obesity in an African population, the first genome-wide linkage analysis for serum lipids in an African population, the first study of genetic structure in West Africans using genome-wide markers and the first genome-wide association study (GWAS) for hypertension and blood pressure in African Americans.
Dr. Adeyemo is currently co-chair of the H3Africa Genome Analysis Working Group and serves on the H3ABioNet Scientific Advisory Board. He is a co-creator of the NHGRI electronic atlas of birth defects for diverse populations.
Scientific Summary
Dr. Adeyemo's research is on the genetic epidemiology of complex diseases within two broad areas: the genetic epidemiology of cardiometabolic disorders and the genetics of complex disease in childhood.
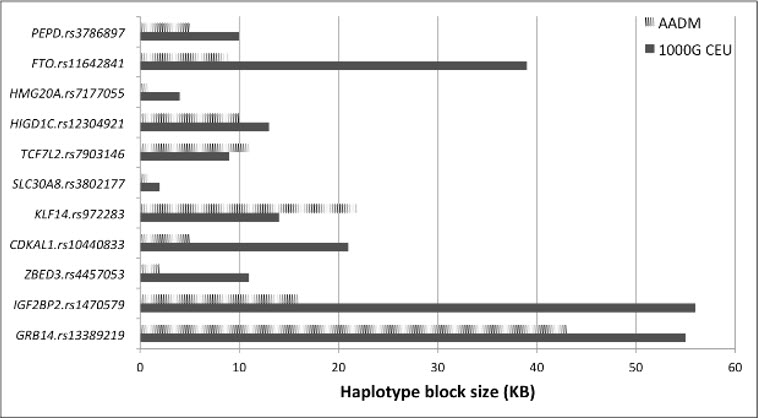
Cardiometabolic disorders, in particular hypertension, obesity and diabetes, are non-communicable conditions of major public health and clinical significance. Dr. Adeyemo's research has focused on the genetic risk factors for these disorders in populations of African ancestry (African Americans and Sub Saharan Africans). Starting with linkage studies (in which he conducted the first genome wide linkage studies for obesity and for lipids in an African population), his studies have progressed through candidate gene studies to genome wide association studies and sequencing studies. Recently, he led the genome wide analysis for type 2 diabetes in the Africa America Diabetes Mellitus (AADM) Study based on genotypes generated from a conventional tag SNP array (the Affymetrix Axiom PanAFR array) and the Affymetrix Axiom Exome Array. Using a sample of ~1800 subjects, the study demonstrated that TCF7L2 showed the strongest association with type 2 diabetes. The study also showed transferability for 41 of 103 tested loci and demonstrated fine mapping of most of the replicated loci, thereby confirming the utility of studying African ancestry populations in such research initiatives. This dataset is currently being combined with another study for the first meta-analysis of type 2 diabetes in Africa.
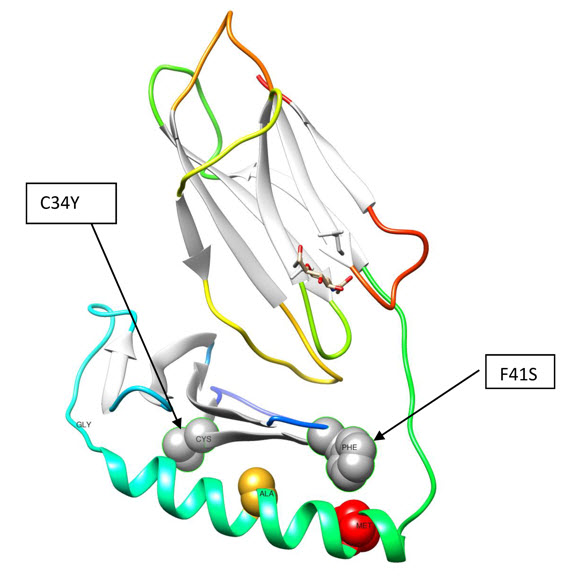
The genomics of complex diseases of childhood has been a research focus for Dr. Adeyemo, bringing together his training and experience in pediatrics, genetics and genetic epidemiology. In collaborative studies, he reported a novel locus (TNXB) for vesico-ureteric reflux (VUR) using a combination of linkage analysis, exome sequencing (WES) and functional studies in a 27-member pedigree (PMID 23620400).
More recently, novel candidate loci (HLADQA1 and PLCG2) for steroid-sensitive nephrotic syndrome (PMID 25349203) were reported in his collaborative studies. This is significant because this was the first time genetic risk variants have been reported for the common form of the nephrotic syndrome, which is the second leading cause of kidney disease in childhood and a major cause of end stage kidney disease requiring dialysis and kidney transplantation in children.
Other studies include: finding that ANLN mutations can cause focal segmental glomerulosclerosis (FSGS) - PMID 24676636; showing that subjects with Alport syndrome with the characteristic mutations in COL4A3/COL4A4 are often misdiagnosed as familial FSGS (PMID 25229338) and characterizing TNXB mutations in a large collection of families with VUR (PMID 26408188).
His research also includes genetics of orofacial clefts (i.e. cleft lip/palate). In collaborative studies, his research led to findings of novel IRF6 mutations in families with Van Der Woude syndrome and popliteal pterygium syndrome in Africa (PMID 24936515) and helped identify rare functional variants in non-syndromic cleft lip/palate (PMID 25081408). Current studies are using a GWAS of ~3000 orofacial clefts patients and controls to identify genetic risk factors for cleft lip/palate in Africa. More recently, his research has grown to include genetics of congenital heart defects. This collaborative study is currently enrolling children with CHD and their parents in Nigeria for genomic studies, including chromosomal arrays and whole exome sequencing. The project is also facilitating the collection of a rich clinical epidemiology dataset of congenital heart defects in an African population
-
Scientific Summary
Dr. Adeyemo's research is on the genetic epidemiology of complex diseases within two broad areas: the genetic epidemiology of cardiometabolic disorders and the genetics of complex disease in childhood.
Cardiometabolic disorders, in particular hypertension, obesity and diabetes, are non-communicable conditions of major public health and clinical significance. Dr. Adeyemo's research has focused on the genetic risk factors for these disorders in populations of African ancestry (African Americans and Sub Saharan Africans). Starting with linkage studies (in which he conducted the first genome wide linkage studies for obesity and for lipids in an African population), his studies have progressed through candidate gene studies to genome wide association studies and sequencing studies. Recently, he led the genome wide analysis for type 2 diabetes in the Africa America Diabetes Mellitus (AADM) Study based on genotypes generated from a conventional tag SNP array (the Affymetrix Axiom PanAFR array) and the Affymetrix Axiom Exome Array. Using a sample of ~1800 subjects, the study demonstrated that TCF7L2 showed the strongest association with type 2 diabetes. The study also showed transferability for 41 of 103 tested loci and demonstrated fine mapping of most of the replicated loci, thereby confirming the utility of studying African ancestry populations in such research initiatives. This dataset is currently being combined with another study for the first meta-analysis of type 2 diabetes in Africa.

The genomics of complex diseases of childhood has been a research focus for Dr. Adeyemo, bringing together his training and experience in pediatrics, genetics and genetic epidemiology. In collaborative studies, he reported a novel locus (TNXB) for vesico-ureteric reflux (VUR) using a combination of linkage analysis, exome sequencing (WES) and functional studies in a 27-member pedigree (PMID 23620400).
More recently, novel candidate loci (HLADQA1 and PLCG2) for steroid-sensitive nephrotic syndrome (PMID 25349203) were reported in his collaborative studies. This is significant because this was the first time genetic risk variants have been reported for the common form of the nephrotic syndrome, which is the second leading cause of kidney disease in childhood and a major cause of end stage kidney disease requiring dialysis and kidney transplantation in children.
Other studies include: finding that ANLN mutations can cause focal segmental glomerulosclerosis (FSGS) - PMID 24676636; showing that subjects with Alport syndrome with the characteristic mutations in COL4A3/COL4A4 are often misdiagnosed as familial FSGS (PMID 25229338) and characterizing TNXB mutations in a large collection of families with VUR (PMID 26408188).

His research also includes genetics of orofacial clefts (i.e. cleft lip/palate). In collaborative studies, his research led to findings of novel IRF6 mutations in families with Van Der Woude syndrome and popliteal pterygium syndrome in Africa (PMID 24936515) and helped identify rare functional variants in non-syndromic cleft lip/palate (PMID 25081408). Current studies are using a GWAS of ~3000 orofacial clefts patients and controls to identify genetic risk factors for cleft lip/palate in Africa. More recently, his research has grown to include genetics of congenital heart defects. This collaborative study is currently enrolling children with CHD and their parents in Nigeria for genomic studies, including chromosomal arrays and whole exome sequencing. The project is also facilitating the collection of a rich clinical epidemiology dataset of congenital heart defects in an African population
Publications
Adeyemo AA, Okolo CM, Omotade OO. Major congenital malformations among paediatric admissions at University College Hospital, Ibadan, Nigeria. Ann Trop Paediatr, 14(1):75-9.1994. [PubMed]
Adeyemo AA, Omotade OO. Variation in fontanelle size with gestational age. Early Hum Dev, 54(3):207-14. 1999. [PubMed]
Adeyemo AA, Prewitt TE, Luke A, Omotade OO, Rotimi CN, Brieger WR, Cooper RS. The feasibility of implementing a dietary sodium reduction intervention among free-living normotensive individuals in south west Nigeria. Ethn Dis, 12(2):207-12. 2002. [PubMed]
Adeyemo AA, Omotade OO, Rotimi CN, Luke AH, Tayo BO, Cooper RS. Heritability of blood pressure in Nigerian families. J Hypertens, 20(5):859-63. 2002. [PubMed]
Adeyemo A, Luke A, Cooper R, Wu X, Tayo B, Zhu X, Rotimi C, Bouzekri N, Ward R. A genome-wide scan for body mass index among Nigerian families. Obes Res, 11(2):266-73. 2003. [PubMed]
Rotimi CN, Chen G, Adeyemo AA, Furbert-Harris P, Parish-Gause D, Zhou J, Berg K, Adegoke O, Amoah A, Owusu S, Acheampong J, Agyenim-Boateng K, Eghan BA Jr, Oli J, Okafor G, Ofoegbu E, Osotimehin B, Abbiyesuku F, Johnson T, Rufus T, Fasanmade O, Kittles R, Daniel H, Chen Y, Dunston G, Collins FS; Africa America Diabetes Mellitus (AADM) Study. A genome-wide search for type 2 diabetes susceptibility genes in West Africans: the Africa America Diabetes Mellitus (AADM) Study. Diabetes, 53(3):838-41. 2004. [PubMed]
Adeyemo AA, Chen G, Chen Y, Rotimi C. Genetic structure in four West African population groups. BMC Genet, 6:38. 2005. [PubMed]
Adeyemo A, Luke A, Wu X, Cooper RS, Kan D, Omotade O, Zhu X. Genetic effects on blood pressure localized to chromosomes 6 and 7. J Hypertens, 23(7):1367-73. 2005. [PubMed]
Adeyemo AA, Johnson T, Acheampong J, Oli J, Okafor G, Amoah A, Owusu S, Agyenim-Boateng K, Eghan BA Jr, Abbiyesuku F, Fasanmade O, Rufus T, Doumatey A, Chen G, Zhou J, Chen Y, Furbert-Harris P, Dunston G, Collins F, Rotimi C. A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis, 181(2):389-97. 2005. [PubMed]
Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics, 13(2):72-9. 2009. [PubMed]
Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet, 5(7):e1000564. 2009. [PubMed]
Chen G, Shriner D, Zhou J, Doumatey A, Huang H, Gerry NP, Herbert A, Christman MF, Chen Y, Dunston GM, Faruque MU, Rotimi CN, Adeyemo A. Development of admixture mapping panels for African Americans from commercial high-density SNP arrays. BMC Genomics, 11:417. 2010. [PubMed]
Adeyemo A, Chen G, Zhou J, Shriner D, Doumatey A, Huang H, Rotimi C. FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes, 59(6):1549-54. 2010. [PubMed]
Faruque MU, Chen G, Doumatey A, Huang H, Zhou J, Dunston GM, Rotimi CN, Adeyemo AA. Association of ATP1B1, RGS5 and SELE polymorphisms with hypertension and blood pressure in African-Americans. J Hypertens, (10):1906-12. 2011. [PubMed]
Tekola Ayele F, Adeyemo A, Finan C, Hailu E, Sinnott P, Burlinson ND, Aseffa A, Rotimi CN, Newport MJ, Davey G. HLA class II locus and susceptibility to podoconiosis. N Engl J Med, 366(13):1200-8. 2012. [PubMed]
Tekola Ayele F, Hailu E, Finan C, Aseffa A, Davey G, Newport MJ, Rotimi CN, Adeyemo A. Prediction of HLA class II alleles using SNPs in an African population. PLoS One, 7(6):e40206. 2012. [PubMed]
Adeyemo A, Bentley AR, Meilleur KG, Doumatey AP, Chen G, Zhou J, Shriner D, Huang H, Herbert A, Gerry NP, Christman MF, Rotimi CN. Transferability and fine mapping of genome-wide associated loci for lipids in African Americans. BMC Med Genet, 13:88. 2012. [PubMed]
Adeyemo A, Tayo BO, Luke A, Ogedegbe O, Durazo-Arvizu R, Cooper RS. The Nigerian antihypertensive adherence trial: a community-based randomized trial. J Hypertens, 31(1):201-7. 2013. [PubMed]
Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, Ritchie GR, Xue Y, Asimit J, Nsubuga RN, Young EH, Pomilla C, Kivinen K, Rockett K, Kamali A, Doumatey AP, Asiki G, Seeley J, Sisay-Joof F, Jallow M, Tollman S, Mekonnen E, Ekong R, Oljira T, Bradman N, Bojang K, Ramsay M, Adeyemo A, Bekele E, Motala A, Norris SA, Pirie F, Kaleebu P, Kwiatkowski D, Tyler-Smith C, Rotimi C, Zeggini E, Sandhu MS. The African Genome Variation Project shapes medical genetics in Africa. Nature, 517(7534):327-32. 2015. [PubMed]
Gbadegesin RA, Brophy PD, Adeyemo A, Hall G, Gupta IR, Hains D, Bartkowiak B, Rabinovich CE, Chandrasekharappa S, Homstad A, Westreich K, Wu G, Liu Y, Holanda D, Clarke J, Lavin P, Selim A, Miller S, Wiener JS, Ross SS, Foreman J, Rotimi C, Winn MP. TNXB mutations can cause vesicoureteral reflux. J Am Soc Nephrol, 24(8):1313-22. 2013. [PubMed]
Butali A, Mossey P, Adeyemo W, Eshete M, Gaines L, Braimah R, Aregbesola B, Rigdon J, Emeka C, Olutayo J, Ogunlewe O, Ladeinde A, Abate F, Hailu T, Mohammed I, Gravem P, Deribew M, Gesses M, Adeyemo A, Marazita M, Murray J. Rare functional variants in genome-wide association identified candidate genes for nonsyndromic clefts in the African population. Am J Med Genet A, 164A(10):2567-71. 2014. [PubMed]
Butali A, Mossey P, Tiffin N, Adeyemo W, Eshete M, Mumena C, Audu R, Onwuamah C, Agbenorku P, Ogunlewe M, Adebola A, Olasoji H, Aregbesola B, Braimah R, Oladugba A, Onah I, Adebiyi E, Olaitan P, Abdur-Rahman L, Adeyemo A. Multidisciplinary approach to genomics research in Africa: the AfriCRAN model. Pan Afr Med J, 21:229. 2015. [PubMed]
Adeyemo AA, Tekola-Ayele F, Doumatey AP, Bentley AR, Chen G, Huang H, Zhou J, Shriner D, Fasanmade O, Okafor G, Eghan B Jr, Agyenim-Boateng K, Adeleye J, Balogun W, Elkahloun A, Chandrasekharappa S, Owusu S, Amoah A, Acheampong J, Johnson T, Oli J, Adebamowo C, Collins F, Dunston G, Rotimi CN. Evaluation of Genome Wide Association Study Associated Type 2 Diabetes Susceptibility Loci in Sub Saharan Africans. Front Genet, 6:335. 2015.[PubMed]
Gbadegesin RA, Adeyemo A, Webb NJ, Greenbaum LA, Abeyagunawardena A, Thalgahagoda S, Kale A, Gipson D, Srivastava T, Lin JJ, Chand D, Hunley TE, Brophy PD, Bagga A, Sinha A, Rheault MN, Ghali J, Nicholls K, Abraham E, Janjua HS, Omoloja A, Barletta GM, Cai Y, Milford DD, O'Brien C, Awan A, Belostotsky V, Smoyer WE, Homstad A, Hall G, Wu G, Nagaraj S, Wigfall D, Foreman J, Winn MP; Mid-West Pediatric Nephrology Consortium. HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol, 26(7):1701-10. 2015. [PubMed]
Elahi S, Homstad A, Vaidya H, Stout J, Hall G, Wu G, Conlon P Jr, Routh JC, Wiener JS, Ross SS, Nagaraj S, Wigfall D, Foreman J, Adeyemo A, Gupta IR, Brophy PD, Rabinovich CE, Gbadegesin RA. Rare variants in tenascin genes in a cohort of children with primary vesicoureteric reflux. Pediatr Nephrol, 31(2):247-53. 2016. [PubMed]
Muenke M, Adeyemo A, Kruszka P. An electronic atlas of human malformation syndromes in diverse populations. Genet Med, 18(11):1085-1087. 2016. [PubMed]
Koretzky M, Bonham VL, Berkman BE, Kruszka P, Adeyemo A, Muenke M, Hull SC. Towards a more representative morphology: clinical and ethical considerations for including diverse populations in diagnostic genetic atlases. Genet Med, 18(11):1069-1074. 2016. [PubMed]
Ekure EN, Adeyemo AA. Clinical epidemiology and management of congenital heart defects in a developing country. In: Muenke M, Kruszka PS, Sable CA, Belmont JW (Eds). Congenital Heart Disease: Molecular Genetics, Principles of Diagnosis and Treatment. Karger, Basel. 2015. Pp. 46-56
-
Publications
Adeyemo AA, Okolo CM, Omotade OO. Major congenital malformations among paediatric admissions at University College Hospital, Ibadan, Nigeria. Ann Trop Paediatr, 14(1):75-9.1994. [PubMed]
Adeyemo AA, Omotade OO. Variation in fontanelle size with gestational age. Early Hum Dev, 54(3):207-14. 1999. [PubMed]
Adeyemo AA, Prewitt TE, Luke A, Omotade OO, Rotimi CN, Brieger WR, Cooper RS. The feasibility of implementing a dietary sodium reduction intervention among free-living normotensive individuals in south west Nigeria. Ethn Dis, 12(2):207-12. 2002. [PubMed]
Adeyemo AA, Omotade OO, Rotimi CN, Luke AH, Tayo BO, Cooper RS. Heritability of blood pressure in Nigerian families. J Hypertens, 20(5):859-63. 2002. [PubMed]
Adeyemo A, Luke A, Cooper R, Wu X, Tayo B, Zhu X, Rotimi C, Bouzekri N, Ward R. A genome-wide scan for body mass index among Nigerian families. Obes Res, 11(2):266-73. 2003. [PubMed]
Rotimi CN, Chen G, Adeyemo AA, Furbert-Harris P, Parish-Gause D, Zhou J, Berg K, Adegoke O, Amoah A, Owusu S, Acheampong J, Agyenim-Boateng K, Eghan BA Jr, Oli J, Okafor G, Ofoegbu E, Osotimehin B, Abbiyesuku F, Johnson T, Rufus T, Fasanmade O, Kittles R, Daniel H, Chen Y, Dunston G, Collins FS; Africa America Diabetes Mellitus (AADM) Study. A genome-wide search for type 2 diabetes susceptibility genes in West Africans: the Africa America Diabetes Mellitus (AADM) Study. Diabetes, 53(3):838-41. 2004. [PubMed]
Adeyemo AA, Chen G, Chen Y, Rotimi C. Genetic structure in four West African population groups. BMC Genet, 6:38. 2005. [PubMed]
Adeyemo A, Luke A, Wu X, Cooper RS, Kan D, Omotade O, Zhu X. Genetic effects on blood pressure localized to chromosomes 6 and 7. J Hypertens, 23(7):1367-73. 2005. [PubMed]
Adeyemo AA, Johnson T, Acheampong J, Oli J, Okafor G, Amoah A, Owusu S, Agyenim-Boateng K, Eghan BA Jr, Abbiyesuku F, Fasanmade O, Rufus T, Doumatey A, Chen G, Zhou J, Chen Y, Furbert-Harris P, Dunston G, Collins F, Rotimi C. A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis, 181(2):389-97. 2005. [PubMed]
Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics, 13(2):72-9. 2009. [PubMed]
Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet, 5(7):e1000564. 2009. [PubMed]
Chen G, Shriner D, Zhou J, Doumatey A, Huang H, Gerry NP, Herbert A, Christman MF, Chen Y, Dunston GM, Faruque MU, Rotimi CN, Adeyemo A. Development of admixture mapping panels for African Americans from commercial high-density SNP arrays. BMC Genomics, 11:417. 2010. [PubMed]
Adeyemo A, Chen G, Zhou J, Shriner D, Doumatey A, Huang H, Rotimi C. FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes, 59(6):1549-54. 2010. [PubMed]
Faruque MU, Chen G, Doumatey A, Huang H, Zhou J, Dunston GM, Rotimi CN, Adeyemo AA. Association of ATP1B1, RGS5 and SELE polymorphisms with hypertension and blood pressure in African-Americans. J Hypertens, (10):1906-12. 2011. [PubMed]
Tekola Ayele F, Adeyemo A, Finan C, Hailu E, Sinnott P, Burlinson ND, Aseffa A, Rotimi CN, Newport MJ, Davey G. HLA class II locus and susceptibility to podoconiosis. N Engl J Med, 366(13):1200-8. 2012. [PubMed]
Tekola Ayele F, Hailu E, Finan C, Aseffa A, Davey G, Newport MJ, Rotimi CN, Adeyemo A. Prediction of HLA class II alleles using SNPs in an African population. PLoS One, 7(6):e40206. 2012. [PubMed]
Adeyemo A, Bentley AR, Meilleur KG, Doumatey AP, Chen G, Zhou J, Shriner D, Huang H, Herbert A, Gerry NP, Christman MF, Rotimi CN. Transferability and fine mapping of genome-wide associated loci for lipids in African Americans. BMC Med Genet, 13:88. 2012. [PubMed]
Adeyemo A, Tayo BO, Luke A, Ogedegbe O, Durazo-Arvizu R, Cooper RS. The Nigerian antihypertensive adherence trial: a community-based randomized trial. J Hypertens, 31(1):201-7. 2013. [PubMed]
Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, Ritchie GR, Xue Y, Asimit J, Nsubuga RN, Young EH, Pomilla C, Kivinen K, Rockett K, Kamali A, Doumatey AP, Asiki G, Seeley J, Sisay-Joof F, Jallow M, Tollman S, Mekonnen E, Ekong R, Oljira T, Bradman N, Bojang K, Ramsay M, Adeyemo A, Bekele E, Motala A, Norris SA, Pirie F, Kaleebu P, Kwiatkowski D, Tyler-Smith C, Rotimi C, Zeggini E, Sandhu MS. The African Genome Variation Project shapes medical genetics in Africa. Nature, 517(7534):327-32. 2015. [PubMed]
Gbadegesin RA, Brophy PD, Adeyemo A, Hall G, Gupta IR, Hains D, Bartkowiak B, Rabinovich CE, Chandrasekharappa S, Homstad A, Westreich K, Wu G, Liu Y, Holanda D, Clarke J, Lavin P, Selim A, Miller S, Wiener JS, Ross SS, Foreman J, Rotimi C, Winn MP. TNXB mutations can cause vesicoureteral reflux. J Am Soc Nephrol, 24(8):1313-22. 2013. [PubMed]
Butali A, Mossey P, Adeyemo W, Eshete M, Gaines L, Braimah R, Aregbesola B, Rigdon J, Emeka C, Olutayo J, Ogunlewe O, Ladeinde A, Abate F, Hailu T, Mohammed I, Gravem P, Deribew M, Gesses M, Adeyemo A, Marazita M, Murray J. Rare functional variants in genome-wide association identified candidate genes for nonsyndromic clefts in the African population. Am J Med Genet A, 164A(10):2567-71. 2014. [PubMed]
Butali A, Mossey P, Tiffin N, Adeyemo W, Eshete M, Mumena C, Audu R, Onwuamah C, Agbenorku P, Ogunlewe M, Adebola A, Olasoji H, Aregbesola B, Braimah R, Oladugba A, Onah I, Adebiyi E, Olaitan P, Abdur-Rahman L, Adeyemo A. Multidisciplinary approach to genomics research in Africa: the AfriCRAN model. Pan Afr Med J, 21:229. 2015. [PubMed]
Adeyemo AA, Tekola-Ayele F, Doumatey AP, Bentley AR, Chen G, Huang H, Zhou J, Shriner D, Fasanmade O, Okafor G, Eghan B Jr, Agyenim-Boateng K, Adeleye J, Balogun W, Elkahloun A, Chandrasekharappa S, Owusu S, Amoah A, Acheampong J, Johnson T, Oli J, Adebamowo C, Collins F, Dunston G, Rotimi CN. Evaluation of Genome Wide Association Study Associated Type 2 Diabetes Susceptibility Loci in Sub Saharan Africans. Front Genet, 6:335. 2015.[PubMed]
Gbadegesin RA, Adeyemo A, Webb NJ, Greenbaum LA, Abeyagunawardena A, Thalgahagoda S, Kale A, Gipson D, Srivastava T, Lin JJ, Chand D, Hunley TE, Brophy PD, Bagga A, Sinha A, Rheault MN, Ghali J, Nicholls K, Abraham E, Janjua HS, Omoloja A, Barletta GM, Cai Y, Milford DD, O'Brien C, Awan A, Belostotsky V, Smoyer WE, Homstad A, Hall G, Wu G, Nagaraj S, Wigfall D, Foreman J, Winn MP; Mid-West Pediatric Nephrology Consortium. HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol, 26(7):1701-10. 2015. [PubMed]
Elahi S, Homstad A, Vaidya H, Stout J, Hall G, Wu G, Conlon P Jr, Routh JC, Wiener JS, Ross SS, Nagaraj S, Wigfall D, Foreman J, Adeyemo A, Gupta IR, Brophy PD, Rabinovich CE, Gbadegesin RA. Rare variants in tenascin genes in a cohort of children with primary vesicoureteric reflux. Pediatr Nephrol, 31(2):247-53. 2016. [PubMed]
Muenke M, Adeyemo A, Kruszka P. An electronic atlas of human malformation syndromes in diverse populations. Genet Med, 18(11):1085-1087. 2016. [PubMed]
Koretzky M, Bonham VL, Berkman BE, Kruszka P, Adeyemo A, Muenke M, Hull SC. Towards a more representative morphology: clinical and ethical considerations for including diverse populations in diagnostic genetic atlases. Genet Med, 18(11):1069-1074. 2016. [PubMed]
Ekure EN, Adeyemo AA. Clinical epidemiology and management of congenital heart defects in a developing country. In: Muenke M, Kruszka PS, Sable CA, Belmont JW (Eds). Congenital Heart Disease: Molecular Genetics, Principles of Diagnosis and Treatment. Karger, Basel. 2015. Pp. 46-56
Last updated: June 21, 2023


