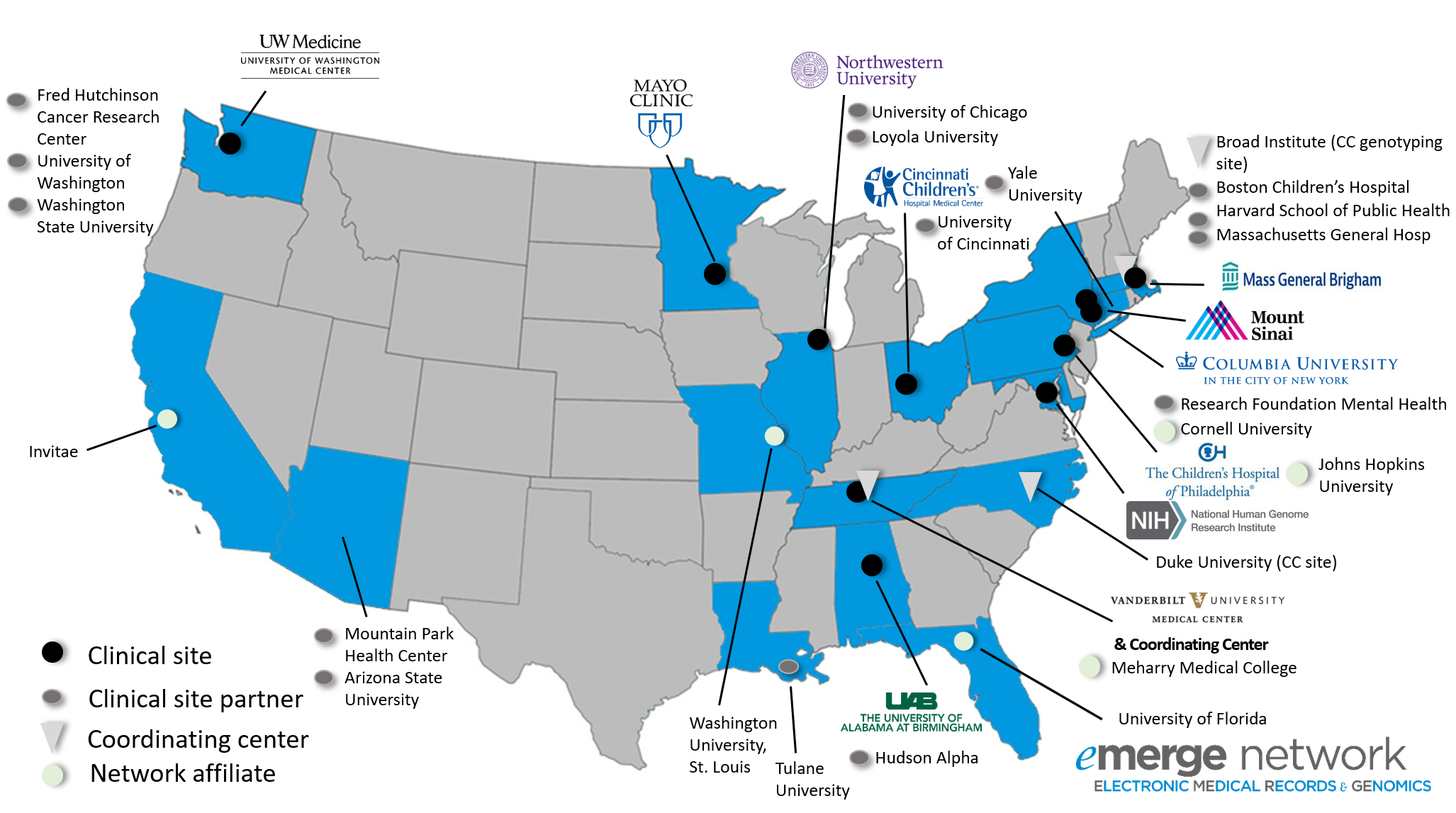
Current Network Participants
Workgroups and Missions
| Working Group | Co-Chairs | Goals |
|---|---|---|
| Comprehensive Risk Assessment and Return | Gail Jarvik, Iftikhar Kullo, and Cindy Prows |
|
| EHR Workflow & Infrastructure | Robert Freimuth and Luke Rasmussen |
|
| Phenotyping | Wei-Qi Wei and Chunhua Weng |
|
| Provider Uptake & Outcomes | Nita Limdi David Veenstra |
|
| PRS Validation & Evaluation | Eimear Kenny Leah Kottyan Niall Lennon |
|
| Recruitment, Retention, sIRB & ELSI | Wendy Chung, Ingrid Holm, and Digna Velez-Edwards |
|
Staff
Program Directors

Robb Rowley, M.D.
- Program Director
- Division of Genomic Medicine

Rene Sterling, Ph.D., M.H.A.
- Program Director
- Division of Genomics and Society
Program Staff

Jessica Reinach, B.A.
- Scientific Program Analyst
- Division of Genomic Medicine
Last updated: March 10, 2025
