Paul P. Liu, M.D., Ph.D.
Senior Investigator
Translational and Functional Genomics Branch
Head
Oncogenesis and Development Section
Education
M.D. Capital Institute of Medicine, Beijing, China
Ph.D. University of Texas
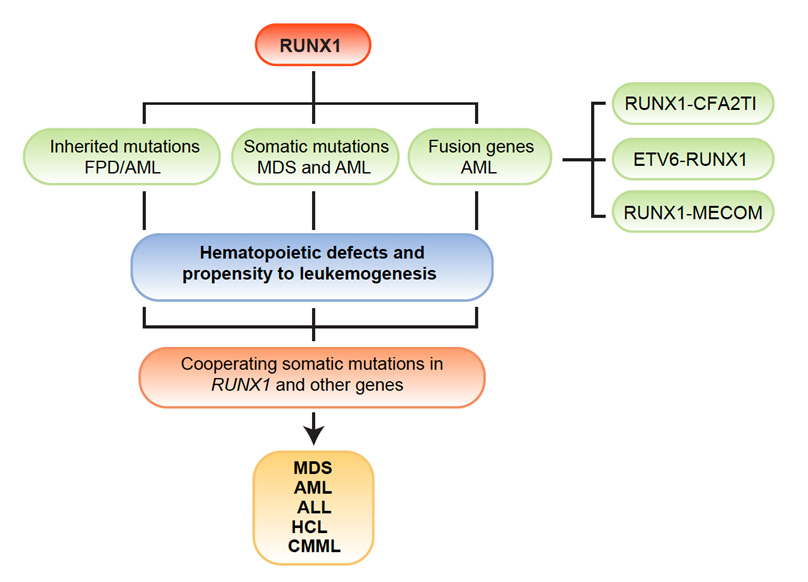
Featured Clinical Study
Oncogenesis and Development Section Staff

Erica Bresciani, Ph.D.
- Staff Scientist
- Oncogenesis and Development Section

Tao Zhen, Ph.D.
- Staff Scientist
- Oncogenesis and Development Section

Natalie Deuitch, M.S., C.G.C.
- Genetic Counselor
- Oncogenesis and Development Section

Shawn Chong, PA-C
- Physician Assistant
- Oncogenesis and Development Section

Kathleen (Katie) Craft, B.S.N., R.N.
- Research Nurse Specialist
- Oncogenesis and Development Section

Catarina (Cat) Menezes, Ph.D.
- Postdoctoral Fellow
- NIH RUNX1-FPD Clinical Research Study

Amra Kajdic, B.S.
- Postbaccalaureate Fellow
- Oncogenesis and Development Section

Aidan D. Pintuff, B.S.
- Postbaccalaureate Fellow
- Oncogenesis and Development Section
Last updated: March 31, 2025