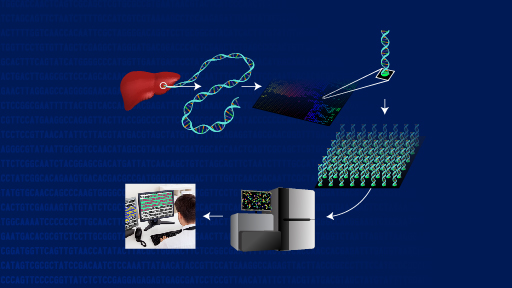
The findings eventually may help guide individualized patient treatment or enrollment in clinical trials that test a therapy aimed at a particular genomic alteration. The scientists reported their results May 18, 2014, in the journal Nature Medicine. Their work is supported by the Clinical Sequencing Exploratory Research program at the National Human Genome Research Institute (NHGRI), and the National Cancer Institute, both part of the National Institutes of Health, in addition to other organizations.
Investigators at Dana-Farber Cancer Institute in Boston, the Broad Institute in Cambridge, Mass., and elsewhere focused on the problem of how the clinician might best use the large amounts of data in a cancer patient's protein-coding exome, the portion of the genome where many key cancer-related genes are located. Initially, they showed that two different tumor sample storage methods produced comparable DNA sequencing information - an important step that may enable genomic profiling to become a more accessible tool for clinicians evaluating cancer patients. The researchers then surveyed established cancer databases, studied the medical literature and spoke to experts to develop a database of 121 tumor genomic alterations that could have therapeutic, prognostic and diagnostic implications for cancer patients.
The researchers subsequently devised an algorithm called the Precision Heuristics for Interpreting the Alteration Landscape (PHIAL), which uses information about a patient's genomic alterations and asks a series of questions to determine which of them might be clinically or biologically important. To validate the tool, they tested its use in previous exome sequencing studies of 511 cancer patients. In 80 percent (408/511) of the patients, PHIAL identified more than 1,800 alterations in the 121 genes and detected potentially important but less common genomic alterations in small subsets of individuals. In a separate study, the algorithm identified 29 genes in 16 current patients that might influence clinical decision-making. In one such case, this system helped connect a patient to a clinical trial.
ARTICLE:
Van Allen, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nature Medicine. 2014. [Full Text]
WHO:
Lucia Hindorff, Ph.D., program director in NHGRI's Clinical Sequencing Exploratory Research program, Division of Genomic Medicine
CONTACT:
To schedule an interview, please contact Steven Benowitz, NHGRI, 301-451-8325, or steven.benowitz@nih.gov
NHGRI is one of the 27 institutes and centers at the National Institutes of Health. The NHGRI Extramural Research Program supports grants for research and training and career development at sites nationwide. Additional information about NHGRI can be found at http://www.genome.gov.
NCI leads the National Cancer Program and the NIH effort to dramatically reduce the prevalence of cancer and improve the lives of cancer patients and their families, through research into prevention and cancer biology, the development of new interventions, and the training and mentoring of new researchers. For more information about cancer, please visit the NCI website at www.cancer.gov or call NCI's Cancer Information Service at 1-800-4-CANCER (1-800-422-6237).
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 institutes and centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit http://www.nih.gov