Open Reading Frame
Definition
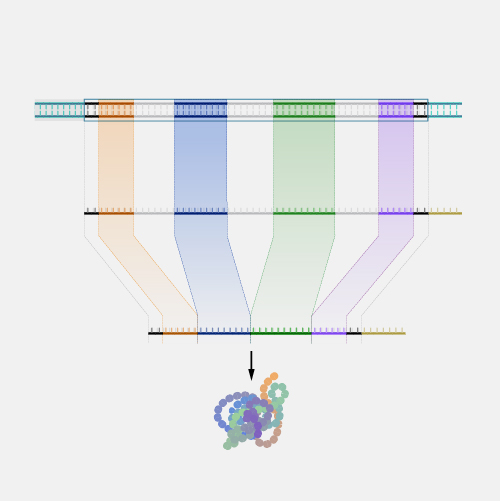
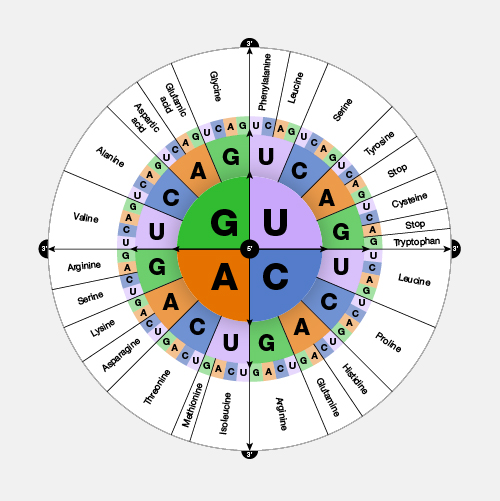
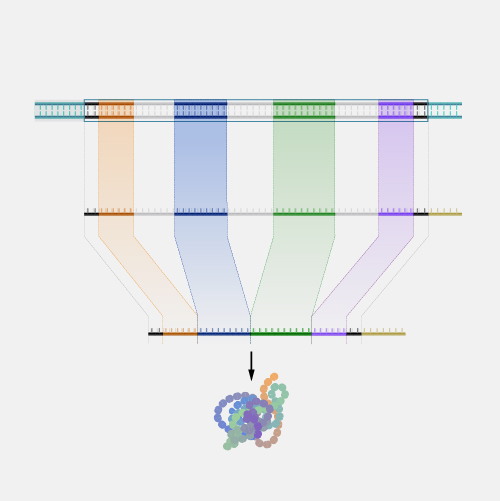
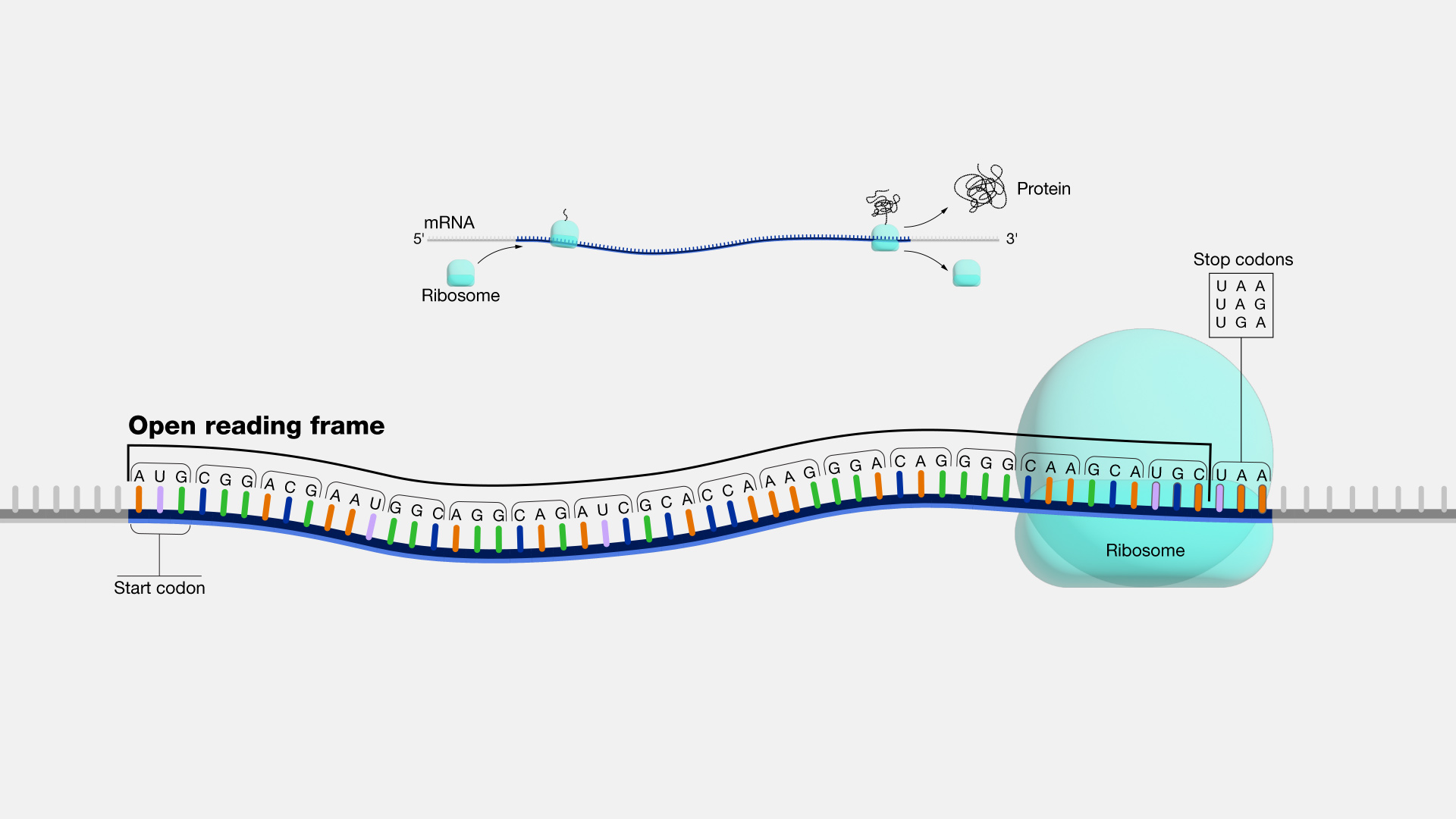
An open reading frame, as related to genomics, is a portion of a DNA sequence that does not include a stop codon (which functions as a stop signal). A codon is a DNA or RNA sequence of three nucleotides (a trinucleotide) that forms a unit of genomic information encoding a particular amino acid or signaling the termination of protein synthesis (stop codon). There are 64 different codons: 61 specify amino acids and 3 are used as stop codons. A long open reading frame is often part of a gene (that is, a sequence directly coding for a protein).
Narration
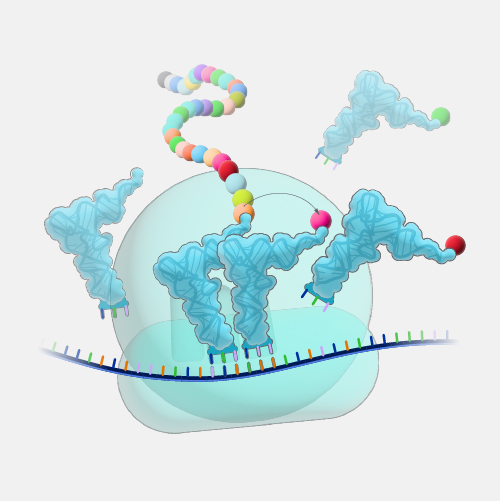
"Open reading frame" is a terrible term that we're stuck with. What it refers to is a frame of reference, and what is being read, "reading", is the RNA code, and it is being read by the ribosomes in order to make a protein. And "open" means that the road is open to keep reading, and the ribosome will be able to keep reading the RNA code and add another amino acid one after another. Now, DNA, though it is a monotonous repetition of As, Cs, Ts, and Gs, has a language, which is transcribed, of course, into RNA and then translated into a protein. And when it's translated into a protein, the mRNA is not read one letter at a time, but it's read three letters at a time. And those three letters are called a codon, and each of those codons, whether it's an AAA or UUU or an AUG, each of those codons is interpreted by the ribosome, the molecular machine, that's going to make the protein as a certain amino acid. So AUG codes for one amino acid, and UUU codes for another, and etc. So an open reading frame is the length of DNA, or RNA, which is transcribed into RNA, through which the ribosome can travel, adding one amino acid after another before it runs into a codon that doesn't code for any amino acid. And when that happens, it confuses the ribosome, and the ribosome stops. So you'll be pleased to hear that codons, which make that happen are called stop codons, and a stop codon ends an open reading frame. So an open reading frame is sometimes 300 amino acids long, and sometimes maybe it's 600, and sometimes it's longer. The longer an open reading frame is, the longer you get before you get to a stop codon, the more likely it is to be part of a gene which is coding for a protein. Now the finally confusing thing about an open reading frame is that because the codons are three nucleic acids long and DNA has two strands, the ribosome can read an RNA derived from one strand or another, and it can read it in 1-2-3s that are separated one from another so you can actually get three reading frames reading in one direction, three reading frames going in the other direction. So it's actually six different reading frames for every piece of DNA, which might give you an open reading frame.