The Big Picture
- Genomic data science is a field of study that enables researchers to use powerful computational and statistical methods to decode the functional information hidden in DNA sequences.
- Estimates predict that genomics research will generate between 2 and 40 exabytes of data within the next decade.
- Our ability to sequence DNA has far outpaced our ability to decipher the information it contains, so genomic data science will be a vibrant field of research for many years to come.
- Performing genomic data science carries with it a set of ethical responsibilities, as each person's sequence data are associated with issues related to privacy and identity.

Because of the sizeable quantity of complex data associated with human genomes, genomics is now considered a "big data" field.

How do scientists study and use genomic data?
Researchers need special computational and analysis tools to find and interpret biological information hidden within the DNA of each person and also to manage the large volumes of data generated in genomics research projects.
Researchers use software tools called aligners to determine where individual pieces of DNA sequence lie on each part of a reference genome sequence.
Next, "variant callers" identify the places where a given human genome sequence differs from other human genome sequences. These genomic differences come in many sizes. The difference may be as small as one DNA letter (called a single-nucleotide polymorphism), many letters long (called structural variants) such as insertions or deletions, or substantially larger chromosomal abnormalities. These genomic differences may present no health risks, or they can directly cause inherited rare disorders, cancers or other more common diseases.

How do researchers manage and store such high volumes of genomic data?
Experts in both computer technologies and genomics manage and store genomic data by using various computer systems and software. Increasingly, data analysis and coordination centers are part of research networks and provide these services.
Generating genomic data requires significant financial support from institutions such as the National Human Genome Research Institute (NHGRI), which provides over $125 million each year to support various genomic data science endeavors.
The generated data resources are often made available to the broader scientific community to facilitate further data analyses. They organize and provide many types of information about the human genome, such as the locations of genes and variants in the DNA.
Many private and commercial cloud platforms work in collaboration with governmental and public entities, such as at the National Institutes of Health (NIH) through the STRIDES initiative. These initiatives provide storage and computing infrastructure for hosting genomic data and to provide the necessary security and privacy protections for human genomic data in particular.
What are some of the ethical, legal and societal implications of genomic data sharing?
Performing genomic research carries with it a set of ethical responsibilities, as information about a person's genome sequence is associated with complex issues related to privacy and identity.
- Informed Consent: Researchers usually ask for consent from individuals whose genomes are sequenced. But researchers must provide clear information about how they will use and share the resulting genome-sequence data in the process of gaining such informed consent.
- Privacy: Powerful computational tools can take sequence data from de-identified genomes and, under special circumstances, connect them back to the person whose DNA was sequenced. Investigators can use such tools for useful purposes, such as identifying criminals who left behind DNA at a crime scene. But the societal benefits must outweigh the potential risks of using genomic data in this way.
- Artificial Intelligence (AI): AI tools increasingly help researchers process vast quantities of genome-sequence data to look for hidden patterns in DNA. However, because AI algorithms often lack transparency, biases can creep in undetected when such algorithms are applied to DNA data. This area of genomic data science will need extensive ethics research to navigate the unique differences between current methods in genomic data science (which rely on human intelligence for interpretation of the results) and newer AI methods. While AI methods offer many promising advantages, they also draw conclusions in completely different ways than humans do, and hence need to be subject to careful ethics oversight.
With all these considerations, data scientists and genomics researchers must be educated about the implications of their studies and work closely with ethics researchers.
How do researchers share human genomic data?
Researchers are expected to share human genomic data according to the consent provided by the research participants. Genomic data are typically shared with the scientific community through data resources, which can be accessed in three ways:
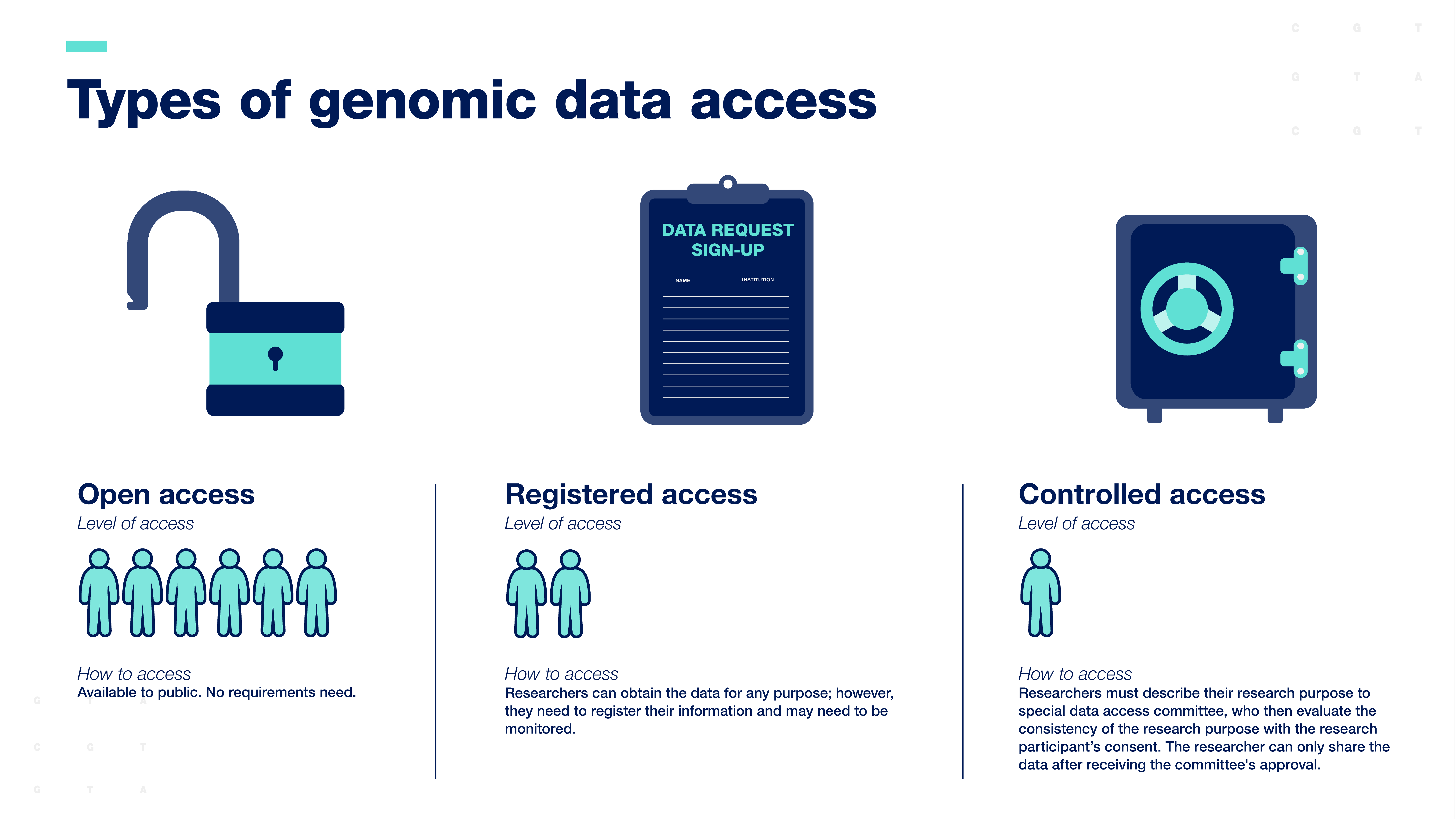
Open-access or unrestricted access is the broadest form of data sharing. Data are available to the public for any research purpose.
Registered access falls in between open-access and controlled-access. Researchers can obtain the data for any purpose; however, they must register their information, and their work with the data may need to be monitored.
Controlled-access data sharing requires researchers to describe their research purpose so that a special data access committee can evaluate the consistency of the research purpose with the participant’s consent. The researcher can only access the data after receiving the committee's approval.

What are some emerging topics in genomic data science?
Human genomes contain many genomic variants (DNA letters that differ in particular places among individuals). Healthcare systems and researchers are building tools that identify these DNA differences and link them to medically relevant information, such as a risk for disease or an indication for a specific medication among several options. Researchers also use artificial intelligence systems to interpret genomic data for clinical purposes, such as diagnosing diseases at early stages or predicting risk for different diseases using genomic information.
In the last decade, cloud computing has become necessary for genomic data storage and analyses. Cloud computing decreases the need to duplicate large datasets, increases security and provides researchers more accessibility to genomic data science. Data scientists are creating tools to make data upload easier and to ensure privacy.

What is NHGRI’s role in working towards a more diverse and equitable workforce?
Women and various minorities groups have been largely underrepresented in genomic data science. NHGRI believes that it is critical to expand and enhance the diversity of the genomic data science workforce. Recent analyses show a significant lack of ethnic and gender diversity among data scientists, trainees and genomics researchers across US institutions.
NHGRI is making changes to enhance the presence of women and underrepresented group in the genomic data science workforce. Through NHGRI-funded training programs and by bringing larger numbers of people in the field of genomic data science, the Institute hopes to foster a more inclusive data science workforce in genomics and beyond.
Additional Resources
To learn more about NHGRI's involvement in genomic data science and data science activities at NIH, see the following resources:
Last updated: April 5, 2022