Genomic Data Sharing Spotlight
In 2022, The Genomics Landscape (TGL) spotlighted genomic data sharing each month in a series of entries. This page is a collection of each of those entries.
On this Page
- Introduction
- The GTEx Dataset
- AnVIL
- Informed Consent for Genomic Data Sharing
- The FAIR and CARE Principles
- Interoperability
- Highly Requested Datasets from the NHGRI Intramural Program
- The Data Sharing Legacy of the Human Genome Project
- The Global Alliance for Genomics and Health
- The FHIR Standard for Clinical Data
- The Upcoming NIH Data Management and Sharing Policy
- Challenges and Opportunities Associated with Genomic Data Sharing
Introduction
From The Genomics Landscape
Originally published: January 13, 2022
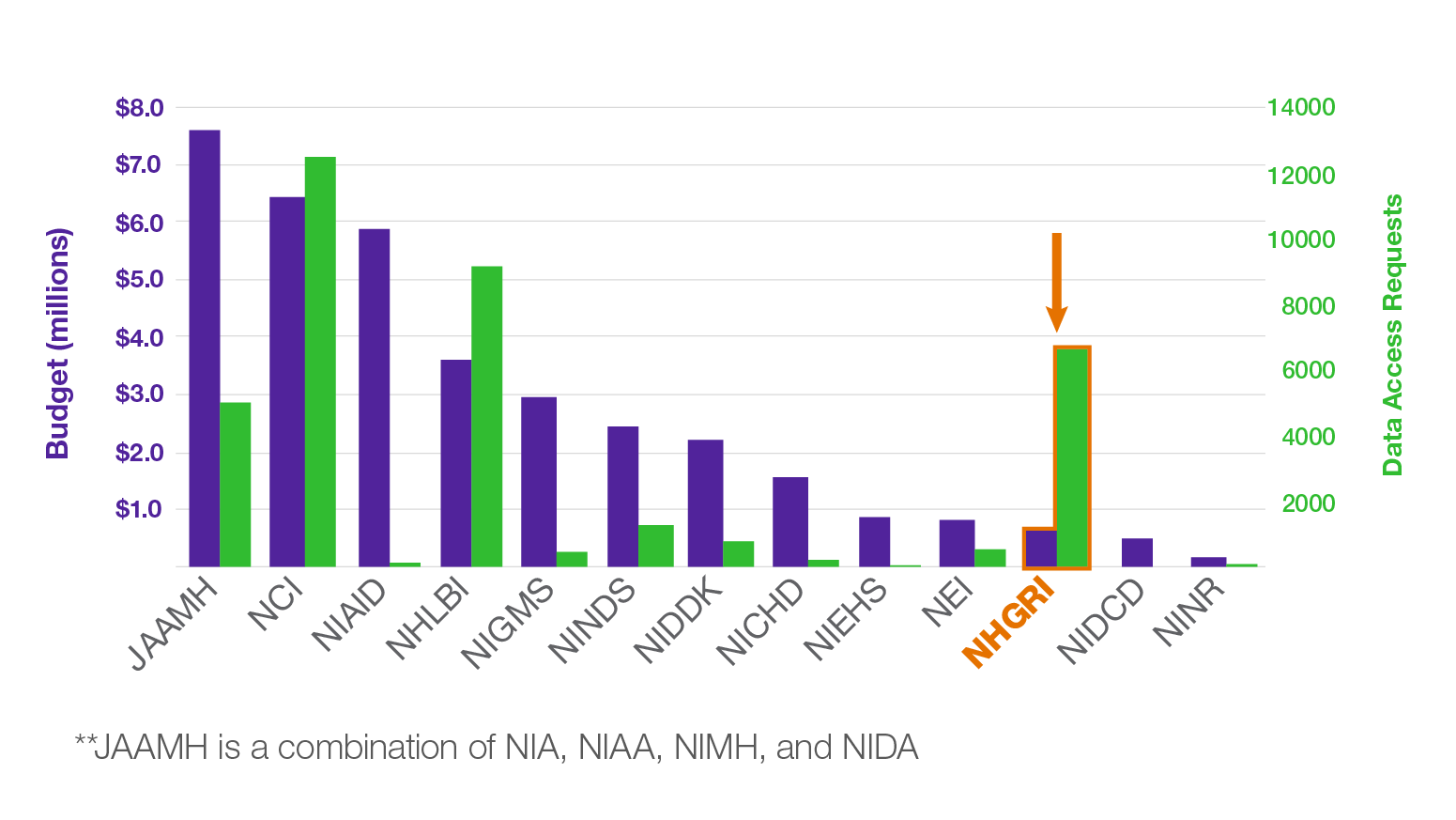
Data sharing is central to the field of genomics. Both human and non-human genomic data have been widely shared through public data resources, such as the database of Genotypes and Phenotypes (dbGaP), model organism databases (e.g., Mouse Genome Informatics and Xenbase), and cloud repositories [e.g., the NHGRI Analysis, Visualization, and Informatics Lab-space (AnVIL)], among others. Data sharing amplifies the impact of research participants’ specimens, increases the power of scientific investigation by allowing genomic data to be combined across studies, and accelerates the pace of discovery by increasing the impact of a single dataset. Over the next year, NHGRI will highlight some interesting facts and stories about how our research community has been at the forefront of data sharing. For example, did you know that although NHGRI is one of the smallest NIH institutes by budget, it ranks #3 in total number of Data Access Requests (DARs) for secondary use of genomic data (see figure)?
The GTEx Dataset
From The Genomics Landscape
Originally published: February 3, 2022
The Genotype-Tissue Expression (GTEx) project generated NHGRI’s most frequently requested controlled-access datasets. It is one of the most widely used resources for studying the relationship between genomic variation and gene expression. Because GTEx data can be shared for general research use, they are being used for many different research purposes. For example, GTEx data are used as controls to study COVID-19 as well as many other diseases and conditions. These datasets are also used to develop computational methods. The GTEx Consortium consists of over 200 investigators from over 40 institutions in the United States and abroad.
The latest version of the GTEx dataset, v8, is now available on the NHGRI cloud repository: NHGRI Analysis Visualization and Informatics Lab-space (AnVIL). Having the data reside in the cloud and running analyses on them there lowers the barrier to performing large-scale computational research by eliminating the need to download data to local platforms. Cloud storage also enhances and enables collaborative efforts. The net effect is an impressive demand for accessing the data; for example, NHGRI received 1237 requests for GTEx controlled-access data in 2020, with 1068 (86%) of those requests approved. Disapprovals were most often related to administrative issues, with most of those requests approved upon resubmission.
AnVIL
From The Genomics Landscape
Originally published: March 7, 2022
In 2018, the NHGRI funded the creation of the Genomic Data Science Analysis, Visualization, and Informatics Lab-space (AnVIL) through cooperative agreements to the Broad Institute and John Hopkins University. AnVIL provides scalable infrastructure, tools, workflows, and training and educational opportunities to facilitate diverse and large-scale genomic-based analyses in a cloud environment. By inverting the traditional model of downloading data, AnVIL eliminates the need for a siloed computing infrastructure and instead makes data accessible in the cloud for users everywhere. Thus, it democratizes access to data for a wider group of researchers and lowers the barrier for large-scale computational research.
In four years, AnVIL has onboarded 4 petabytes of data. That is the equivalent of over 26 billion photos on Facebook. Major research programs whose data are now available in AnVIL via open or controlled access are: 1,000 Genomes, Centers for Common Disease Genomics, Centers for Mendelian Genomics, Whole Genome Sequencing in Psychiatric Disorders, Convergent Neuroscience, the Genotype-Tissue Expression project, Human Pangenome Reference Consortium, Population Architecture Using Genomics and Epidemiology Consortium, and the Telomere-to-Telomere consortium. In addition, AnVIL has recently integrated seqr (an open-source, web-based platform for family-based monogenic disease analysis) into its suite of tools, which also includes familiar tools and workflows, such as those in Bioconductor, Dockstore, Jupyter, and Galaxy.
Are you accessing AnVIL resources for your research? We want to hear from you. Please share recent publications, plans, and innovative workflows with nhgrigds@nih.gov. You may see your research highlighted in a future issue of this newsletter!

Informed Consent for Genomic Data Sharing
From The Genomics Landscape
Originally published: April 7, 2022
When pursuing clinical genomic studies, it is important to be transparent with both research participants and patients about how their genomic data will be used and shared. In the clinical setting, where patients are not necessarily enrolled in a research study when first seeking clinical care, conversations about how genomic data may later be shared in databases can sometimes be an afterthought, making it difficult to determine whether and how data can be shared down the line. To address this issue, the ClinGen Education, Communication, and Training Working Group developed a helpful one page consent form and training materials, which could be used as a model by others in point-of-care settings when collecting specimens that may later be used to generate genomic data that will be submitted to a database for future clinical and/or research use. Available in four different languages, the document is now accessible to a wide range of patients.
As the lines between patient care and research continue to evolve and blur, these sorts of mechanisms for patient consent are critical to maintaining the trust of the public in genomics research. Did you know that the top contributors of clinically relevant genomic variants to ClinVar are commercial laboratories? These groups are doing their part to ensure that future patients, no matter where they have their genomic tests performed, can have a better picture of the clinical significance of their results.
FAIR and CARE Principles
From The Genomics Landscape
Originally published: May 5, 2022
What are FAIR Principles, and what’s all the fuss? In 2016, stakeholders came together to endorse “a concise and measurable set of principles” to improve the management and sharing of research data. Findable, Accessible, Interoperable, and Reusable (FAIR) principles are critical elements for making data maximally useful to the research community. Not long after, the Collective Benefit, Authority to Control, Responsibility, and Ethics (CARE) Principles for Indigenous Data Governance were developed, which aimed to ensure that data-sharing movements (like FAIR) would also consider Indigenous peoples’ goals, values, and rights to self-determination.
When you are collecting data that must be shared broadly with the research community, are you following FAIR and CARE Principles? Are you consistently recording metadata (i.e., data that provides relevant information about the data), using commonly accepted data standards, submitting to a FAIR data repository, and working responsibly with Tribal individuals and communities? One must consider these aspects up front, so that responsible use of the data is maximized down the road.
Did you know that NHGRI released a notice in 2021 that outlined expectations for sharing comprehensive and standardized metadata and phenotypic data? The upcoming NIH Data Management and Sharing Policy (which will become effective on January 23, 2023) also requires that investigators who apply for NIH funding describe the data they plan to share and the data standards they plan to use. In addition, the draft supplemental information to the NIH Policy for Data Management and Sharing: Responsible Management and Sharing of American Indian/Alaska Native Participant Data provides information about helpful resources to aid researchers working with Tribal communities.

Interoperability
From The Genomics Landscape
Originally published: June 2, 2022
Finding a needle in a haystack is never an easy task. In the field of genomics, this analogy often applies when trying to identify genomic variants associated with phenotypes in large-scale sequence datasets. NIH is increasingly hosting genome-sequence datasets and other genomic datasets in cloud environments. Cloud-based computing helps researchers more efficiently sift through the haystack to find that needle. Pooling datasets from multiple studies enhances the statistical power of the resulting analyses, thereby increasing the likelihood that scientists will find both rare and common genomic variants that contribute to health and disease.
Enabling scientists to seamlessly combine datasets that were generated in different studies and that are housed in different repositories requires well-defined standards for interoperability. To this end, NIH is supporting efforts to improve the interoperability of the emerging set of NIH-supported cloud-based platforms and resources. When researchers access data from one site, there is no guarantee that those data will be readily usable along with data from a different site. By focusing on interoperability, the new NIH effort is improving the ability of researchers to integrate and analyze the data derived from multiple different datasets.
Are you interested in what it takes to build interoperable genomic resources in the cloud? The NIH Cloud Platforms Interoperability (NCPI) Effort will be hosting its first open meeting on June 22 and 23. Did you know that the existing NCPI sites currently house a whopping 11 petabytes of data? As the various genomic resources continue to grow and interoperate, so too will the power of human genomic studies.

Highly Requested Datasets from the NHGRI Intramural Program
From The Genomics Landscape
Originally published: July 7, 2022
As with all NIH-funded work, NHGRI intramural researchers must responsibly share large-scale genomic data with the broader research community. The two most highly requested datasets that have been deposited from NHGRI intramural research groups to date are the FUSION study and the ClinSeq Project.
“The FUSION Study: Mapping Genes for Non-Insulin Dependent Diabetes Mellitus - Tissue Biopsy Study” is a long-term, international, and collaborative effort to identify genomic variants that predispose to type 2 diabetes. Nearly 60 other researchers have used this dataset (phs001048), which includes whole-genome sequence, transcriptomic data, and methylation data. These data were used in studies covering a variety of topics: chronic complex diseases, type 2 diabetes, epigenomics, aging, cancer, data privacy, and others.
The ClinSeq Project is a genomic medicine study to investigate the utility of whole-genome sequencing in the practice of medicine. The ClinSeq dataset (phs000971) features a robust list of clinical data types, which has turned out to be useful for exploring different research questions. A key aim of the ClinSeq Project is to inspire further studies in genomic medicine implementation; thus, this dataset is particularly useful for those seeking to answer clinical genomics questions. In addition, this dataset is also made accessible to NIH intramural researchers for exploration of genotype information through the relatively new Reverse Phenotyping Core in the NHGRI Center for Precision Health Research.
Did you know that there are ~1,000 principal investigators and more than 4,000 trainees at various stages of their career conducting basic, translational, and clinical research at the NIH? This makes the NIH Intramural Research Program the largest biomedical research institution on earth!
The Data Sharing Legacy of the Human Genome Project
From The Genomics Landscape
Originally published: August 4, 2022
Did you know that the goal of researchers contributing to the Human Genome Project (HGP) was to submit their sequence data into a public database within 24 hours of being generated? This target was documented during a strategy meeting in Bermuda in 1997, which is why this expectation is often referred to as the “Bermuda Principles.” The legacy of rapid data sharing within the HGP, as well as other large collaborative genomics projects, remains today. Open, quick, and quality data sharing—though not without its challenges—is an ideal that scientists, funders, journals, and many other stakeholders strive for across the biomedical research landscape. To read more about the origins of the Bermuda Principles, see here.
The NIH, inspired by the success of the HGP and other data-sharing efforts, has been advancing its data-sharing policies over the last two decades. NHGRI, as a leader in genomics, has also developed policies and practices that go beyond the baseline set by NIH. For instance, NHGRI expects researchers to leverage data standards and provide comprehensive metadata and phenotypic data for data shared per NIH data-sharing policies. The upcoming NIH Data Management and Sharing (DMS) Policy makes it more important than ever to ensure that shared data are maximally useful to the broader scientific community. NIH will be holding a webinar series in August and September focused implementing this policy. To learn more about the upcoming policy, see sharing.nih.gov and register now for the two-part interactive series to learn what the DMS policy means for you!

The Global Alliance for Genomics and Health
From The Genomics Landscape
Originally published: September 1, 2022
The Global Alliance for Genomics and Health (GA4GH) is a standards-setting organization for genomics data sharing, similar in principle to the World Wide Web Consortium that helped build the foundations of the internet. GA4GH strives to enable international data sharing by cultivating standards that will make it easier for researchers around the world to find, access, combine, and securely analyze genomic and health-related data. By adhering to similar policy frameworks and technical standards, GA4GH hopes to help researchers reach across time zones and national barriers to drive discoveries in human genomics. GA4GH standards are designed to work in tandem to enable data sharing and analysis from start to finish. The organization welcomes engagement and is developing tools and starter kits that will make adopting the standards easier for newcomers or those with less technical expertise.
GA4GH is fueled by volunteers who are actively improving the genomic data sharing ecosystem. Whether you’re interested in learning about existing GA4GH standards, helping to develop or improve a technical standard, or testing out a standard where you work, we encourage you to reach out to info@ga4gh.org. Did you know that GA4GH contributors hail from more than 60 countries around the world?
To learn more about the mission of GA4GH, check out this short video.
Image by GA4GH
The FHIR Standard for Clinical Data
From The Genomics Landscape
Originally published: October 6, 2022
HL7 Fast Healthcare Interoperability Resources (FHIR) is a clinical data standard and an application programming interface to exchange Electronic Health Records (EHRs). FHIR provides an opportunity for the genomic medicine research community to develop ways to consistently report genomics and genomics-related data among different healthcare systems. However, additional developments are needed to better understand how these data should be represented in FHIR in order to best serve both the clinical and basic research communities. In 2019, NIH issued a notice encouraging researchers to explore the use of FHIR to capture, integrate, and exchange clinical data for research purposes and to enhance capabilities to share research data. Did you know that NHGRI-supported programs are involved in improving FHIR’s ability to exchange genomics and genomics-related information among discrete healthcare systems? For example, the Electronic Medical Records and Genomics (eMERGE) Network has been involved in expanding the capabilities of FHIR to represent clinical genomics results, and, more recently, FHIR is being incorporated into NHGRI resource programs such as the Genomic Data Science Analysis, Visualization, and Informatics Lab-space (AnVIL).
Efforts to standardize genomic medicine research are not finished. In February 2021, the NHGRI Genomic Medicine Working Group hosted a virtual meeting titled “Genomic Medicine XIII: Developing a Clinical Genomic Informatics Research Agenda.” The meeting called out “identifying and addressing semantic and syntactic gaps related to the representation of genomic information in existing clinical data standards and models” as a short-term need for the clinical informatics field. Over the coming years, NHGRI will be focusing on this issue, so as to pave the way for genomics to be responsibly incorporated into the clinical setting.

The Upcoming NIH Data Management and Sharing Policy
From The Genomics Landscape
Originally published November 3, 2022
The highly anticipated new NIH Data Management and Sharing (DMS) Policy will go into effect in just under three months, specifically on January 25, 2023. At that time, NIH-supported researchers generating scientific data will be required to submit a DMS Plan as part of their grant application or project proposal. The goal of the new policy is to promote data management and sharing, thereby enabling validation of research results, providing accessibility to high-value datasets, and promoting data reuse for future research studies. To help the NHGRI community of investigators, the institute updated its website to clarify expectations for both the new policy and the long-standing NIH Genomic Data Sharing Policy; this includes the provision of resources to help investigators navigate the two policies. We encourage interested investigators to check out the new webpage and to send comments to nhgrigds@nih.gov!
NHGRI is committed to working with the genomics research community to maximize the quality of data that are shared based on the two NIH data sharing policies, in addition to helping with the sharing of research software and code. With time, the institute expects that investigators will be more equipped to share the valuable data and other products generated through their work. For example, the sharing of well-annotated data will foster reproducibility and enable the emerging fields of artificial intelligence and machine learning (AI/ML) to transform how we integrate and analyze large datasets, ultimately leading to significant advances in human health and healthcare. In fact, NIH has various initiatives aiming to catalyze AI/ML work, including efforts that have a strong genomics component (learn more).

Challenges and Opportunities Associated with Genomic Data Sharing
From The Genomics Landscape
Originally published: December 8, 2022
Over the past year, various aspects of genomic data sharing (GDS) have been highlighted via this spotlight series. There are many stakeholders that are part of this data-sharing ecosystem – patients and research participants, those that manage data repositories, data-access committees, policymakers, standards-generating organizations, and many more. The initial inspiration for this GDS Spotlight series was a conversation in 2020 that NHGRI hosted with scientific journal editors, another important group of stakeholders. Scientific journals share in the responsibility to ensure that the data underlying published research findings are made public and meet appropriate expectations for scientific rigor and reproducibility. That conversation highlighted several challenges that can complicate data sharing, including consent, data ownership, privacy, and international regulations. By increasing awareness about the importance and nuances of data sharing, the research community will hopefully be proactive in facilitating data sharing from the very beginning of each new study. Did you know that NHGRI maintains an Informed Consent Resource to provide guidance and sample language for genomics research, including how to describe data sharing to research participants?
The entries for this year’s GDS Spotlight series are now available on this page for future reference. Hopefully, others will get involved in this campaign to inform the community about the resources available to both share data and make those data maximally useful down the line!

Last updated: December 8, 2022
