Data Sharing Policies and Expectations
This webpage and the associated FAQs describe the various expectations for data sharing that are specific to NHGRI-supported studies. The NHGRI Data Sharing Governance Committee oversees the institute's implementation and maintenance of NIH and NHGRI data sharing policies. For general NIH data sharing policy information, please visit NIH's Scientific Data Sharing website.
Explore This Page
How to Register Controlled-Access Studies
Study registration in dbGaP is required for large-scale human genomic studies, including those submitting data to AnVIL and studies with an Alternative Data Sharing Plan.
Follow the standard process outlined in How to Register and Submit Your Study in dbGaP (Steps 1 – 6) to register your study in dbGaP. Next, see the AnVIL Submission Guide for instructions on submitting the data (Step 7).
For Step 2: Please complete the relevant template below or send the basic study information needed for study registration to the NHGRI GPA (nhgrigpa@mail.nih.gov):
- NHGRI’s Basic Study Information Template

- Template for Requesting an Exception for Samples Lacking Explicit Consent for Future Research Use and Broad Data Sharing

Investigators seeking to submit non-NIH funded data to an NIH-designated data repository (e.g., AnVIL or dbGaP) should follow these instructions.
Timelines for Submitting and Releasing Data
NHGRI follows the NIH’s expectation for submission and release of scientific data, with the following exception: for genomic data, NHGRI expects non-human genomic data that are subject to the NIH GDS Policy to be submitted and released on the same timeline as human genomic data.
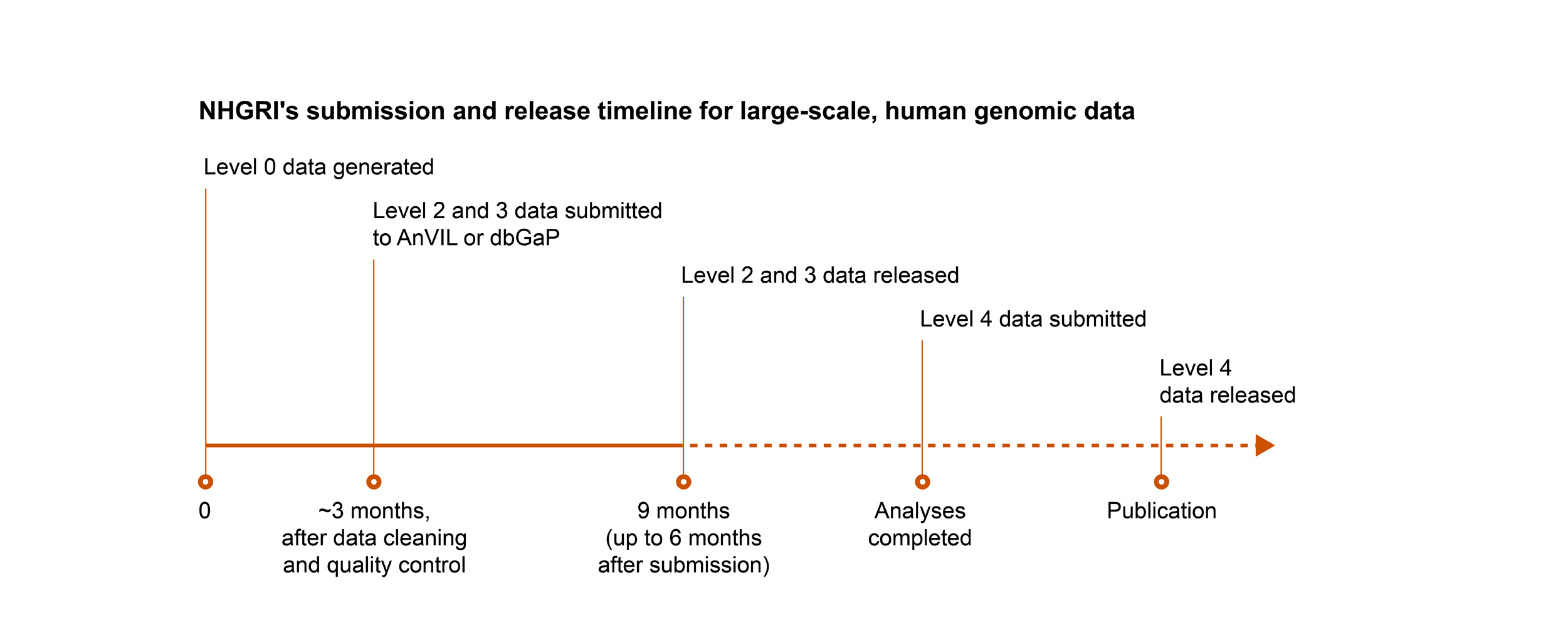
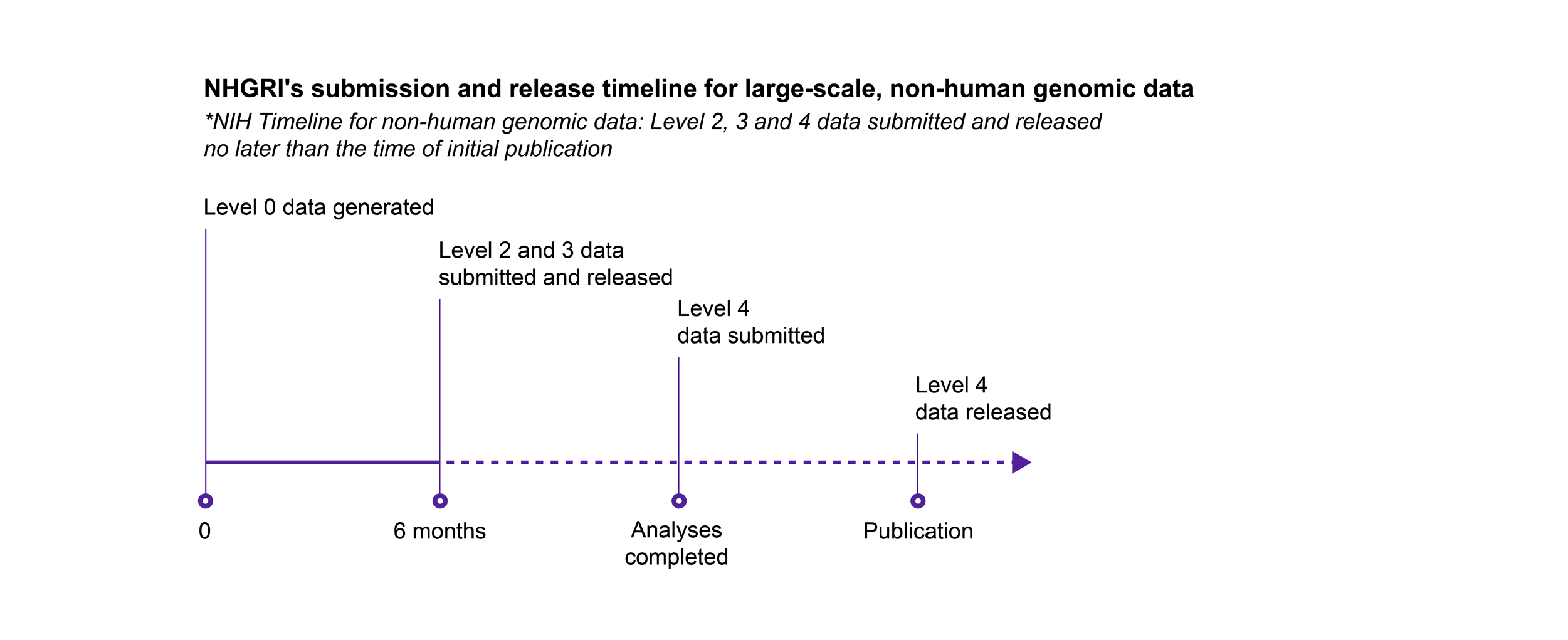
Key
- Level 0: Raw data generated directly from the instrument platform
- Level 1: Initial sequence reads, the most fundamental form of the data after the basic translation of raw input
- Level 2: Data after an initial round of analysis or computation to clean the data and assess basic quality measures
- Level 3: Analysis to identify genetic variants, gene expression patterns, or other features of the data set
- Level 4: Final analysis that relates the genomic data to phenotype or other biological states
Contacts
For Specific Data Sharing Policy Questions:

- NHGRI Data Access Committee (DAC) Chair
- Scientific Review Branch

- Genomic Program Administrator
- National Human Genome Research Institute

- Alternate Genomic Program Administrator
- National Human Genome Research Institute
For Policy Questions:

- Policy Advisor for Data Science and Sharing
- Office of Genomic Data Science
Last updated: February 11, 2025
